Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1
- PMID: 20605055
- PMCID: PMC2958305
- DOI: 10.1016/j.ejcb.2010.05.003
Studies on the morphology and spreading of human endothelial cells define key inter- and intramolecular interactions for talin1
Abstract
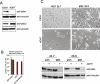
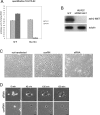
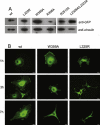
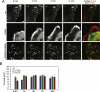
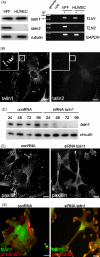
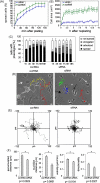
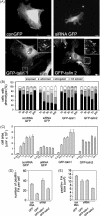
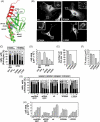
Talin binds to and activates integrins and is thought to couple them to cytoskeletal actin. However, functional studies on talin have been restricted by the fact that most cells express two talin isoforms. Here we show that human umbilical vein endothelial cells (HUVEC) express only talin1, and that talin1 knockdown inhibited focal adhesion (FA) assembly preventing the cells from maintaining a spread morphology, a phenotype that was rescued by GFP-mouse talin1. Thus HUVEC offer an ideal model system in which to conduct talin structure/function studies. Talin contains an N-terminal FERM domain (comprised of F1, F2 and F3 domains) and a C-terminal flexible rod. The F3 FERM domain binds beta-integrin tails, and mutations in F3 that inhibited integrin binding (W359A) or activation (L325R) severely compromised the ability of GFP-talin1 to rescue the talin1 knockdown phenotype despite the presence of a second integrin-binding site in the talin rod. The talin rod contains several actin-binding sites (ABS), and mutations in the C-terminal ABS that reduced actin-binding impaired talin1 function, whereas those that increased binding resulted in more stable FAs. The results show that both the N-terminal integrin and C-terminal actin-binding functions of talin are essential to cell spreading and FA assembly. Finally, mutations that relieve talin auto-inhibition resulted in the rapid and excessive production of FA, highlighting the importance of talin regulation within the cell.
Copyright 2010 Elsevier GmbH. All rights reserved.
Figures
Similar articles
-
Talin contains a C-terminal calpain2 cleavage site important in focal adhesion dynamics.PLoS One. 2012;7(4):e34461. doi: 10.1371/journal.pone.0034461. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496808 Free PMC article.
-
Talin-dependent integrin activation is required for endothelial proliferation and postnatal angiogenesis.Angiogenesis. 2021 Feb;24(1):177-190. doi: 10.1007/s10456-020-09756-4. Epub 2020 Oct 28. Angiogenesis. 2021. PMID: 33113074 Free PMC article.
-
The Architecture of Talin1 Reveals an Autoinhibition Mechanism.Cell. 2019 Sep 19;179(1):120-131.e13. doi: 10.1016/j.cell.2019.08.034. Cell. 2019. PMID: 31539492 Free PMC article.
-
Talin‑1 interaction network in cellular mechanotransduction (Review).Int J Mol Med. 2022 May;49(5):60. doi: 10.3892/ijmm.2022.5116. Epub 2022 Mar 10. Int J Mol Med. 2022. PMID: 35266014 Free PMC article. Review.
-
Integrin-mediated cell adhesion: the cytoskeletal connection.Biochem Soc Symp. 1999;65:79-99. Biochem Soc Symp. 1999. PMID: 10320934 Review.
Cited by
-
FAK promotes recruitment of talin to nascent adhesions to control cell motility.J Cell Biol. 2012 Jan 23;196(2):223-32. doi: 10.1083/jcb.201108078. J Cell Biol. 2012. PMID: 22270917 Free PMC article.
-
Induction of focal adhesions and motility in Drosophila S2 cells.Mol Biol Cell. 2014 Dec 1;25(24):3861-9. doi: 10.1091/mbc.E14-04-0863. Epub 2014 Oct 1. Mol Biol Cell. 2014. PMID: 25273555 Free PMC article.
-
Inorganic polyphosphate induces accelerated tube formation of HUVEC endothelial cells.Cell Mol Life Sci. 2018 Jan;75(1):21-32. doi: 10.1007/s00018-017-2601-2. Epub 2017 Aug 2. Cell Mol Life Sci. 2018. PMID: 28770290 Free PMC article.
-
Over-expressed, N-terminally truncated BRAF is detected in the nucleus of cells with nuclear phosphorylated MEK and ERK.Heliyon. 2018 Dec 20;4(12):e01065. doi: 10.1016/j.heliyon.2018.e01065. eCollection 2018 Dec. Heliyon. 2018. PMID: 30603699 Free PMC article.
-
The Structure of the talin head reveals a novel extended conformation of the FERM domain.Structure. 2010 Oct 13;18(10):1289-99. doi: 10.1016/j.str.2010.07.011. Structure. 2010. PMID: 20947018 Free PMC article.
References
-
- Albiges-Rizo C., Frachet P., Block M.R. Down regulation of talin alters cell adhesion and the processing of the a5b1 integrin. J. Cell Sci. 1995;108:3317–3329. - PubMed
-
- Arias-Salgado E.G., Lizano S., Shattil S.J., Ginsberg M.H. Specification of the direction of adhesive signaling by the integrin beta cytoplasmic domain. J. Biol. Chem. 2005;280:29699–29707. - PubMed
-
- Bolton S.J., Barry S.T., Mosley H., Patel B., Jockusch B., Wilkinson J.M., Critchley D.R. Monoclonal antibodies recognizing the N- and C-terminal regions of talin disrupt actin stress fibers when microinjected into human fibroblasts. Cell Motil. Cytoskeleton. 1997;36:363–376. - PubMed
-
- Bouaouina M., Lad Y., Calderwood D.A. The N-terminal domains of talin cooperate with the phosphotyrosine binding-like domain to activate beta1 and beta3 integrins. J. Biol. Chem. 2008;283:6118–6125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

