Association between the herpes simplex virus-1 DNA polymerase and uracil DNA glycosylase
- PMID: 20601642
- PMCID: PMC2934634
- DOI: 10.1074/jbc.M110.131235
Association between the herpes simplex virus-1 DNA polymerase and uracil DNA glycosylase
Abstract
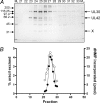
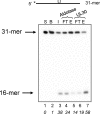
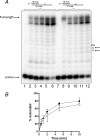
Herpes simplex virus-1 (HSV-1) is a large dsDNA virus that encodes its own DNA replication machinery and other enzymes involved in DNA transactions. We recently reported that the HSV-1 DNA polymerase catalytic subunit (UL30) exhibits apurinic/apyrimidinic and 5'-deoxyribose phosphate lyase activities. Moreover, UL30, in conjunction with the viral uracil DNA glycosylase (UL2), cellular apurinic/apyrimidinic endonuclease, and DNA ligase IIIalpha-XRCC1, performs uracil-initiated base excision repair. Base excision repair is required to maintain genome stability as a means to counter the accumulation of unusual bases and to protect from the loss of DNA bases. Here we show that the HSV-1 UL2 associates with the viral replisome. We identified UL2 as a protein that co-purifies with the DNA polymerase through numerous chromatographic steps, an interaction that was verified by co-immunoprecipitation and direct binding studies. The interaction between UL2 and the DNA polymerase is mediated through the UL30 subunit. Moreover, UL2 co-localizes with UL30 to nuclear viral prereplicative sites. The functional consequence of this interaction is that replication of uracil-containing templates stalls at positions -1 and -2 relative to the template uracil because of the fact that these are converted into non-instructional abasic sites. These findings support the existence of a viral repair complex that may be capable of replication-coupled base excision repair and further highlight the role of DNA repair in the maintenance of the HSV-1 genome.
Figures






Similar articles
-
Reconstitution of uracil DNA glycosylase-initiated base excision repair in herpes simplex virus-1.J Biol Chem. 2009 Jun 19;284(25):16784-16790. doi: 10.1074/jbc.M109.010413. Epub 2009 May 1. J Biol Chem. 2009. PMID: 19411250 Free PMC article.
-
The replicative DNA polymerase of herpes simplex virus 1 exhibits apurinic/apyrimidinic and 5'-deoxyribose phosphate lyase activities.Proc Natl Acad Sci U S A. 2008 Aug 19;105(33):11709-14. doi: 10.1073/pnas.0806375105. Epub 2008 Aug 11. Proc Natl Acad Sci U S A. 2008. PMID: 18695225 Free PMC article.
-
Proliferating cell nuclear antigen inhibitors block distinct stages of herpes simplex virus infection.PLoS Pathog. 2023 Jul 24;19(7):e1011539. doi: 10.1371/journal.ppat.1011539. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37486931 Free PMC article.
-
HSV-1 DNA Replication-Coordinated Regulation by Viral and Cellular Factors.Viruses. 2021 Oct 7;13(10):2015. doi: 10.3390/v13102015. Viruses. 2021. PMID: 34696446 Free PMC article. Review.
-
The vaccinia virus DNA polymerase and its processivity factor.Virus Res. 2017 Apr 15;234:193-206. doi: 10.1016/j.virusres.2017.01.027. Epub 2017 Feb 1. Virus Res. 2017. PMID: 28159613 Free PMC article. Review.
Cited by
-
Distribution and effects of amino acid changes in drug-resistant α and β herpesviruses DNA polymerase.Nucleic Acids Res. 2016 Nov 16;44(20):9530-9554. doi: 10.1093/nar/gkw875. Epub 2016 Sep 29. Nucleic Acids Res. 2016. PMID: 27694307 Free PMC article. Review.
-
The Essential Co-Option of Uracil-DNA Glycosylases by Herpesviruses Invites Novel Antiviral Design.Microorganisms. 2020 Mar 24;8(3):461. doi: 10.3390/microorganisms8030461. Microorganisms. 2020. PMID: 32214054 Free PMC article. Review.
-
Targeting DNA polymerase ß for therapeutic intervention.Curr Mol Pharmacol. 2012 Jan;5(1):68-87. Curr Mol Pharmacol. 2012. PMID: 22122465 Free PMC article. Review.
-
Absence of the uracil DNA glycosylase of murine gammaherpesvirus 68 impairs replication and delays the establishment of latency in vivo.J Virol. 2015 Mar;89(6):3366-79. doi: 10.1128/JVI.03111-14. Epub 2015 Jan 14. J Virol. 2015. PMID: 25589640 Free PMC article.
-
Interaction of the human cytomegalovirus uracil DNA glycosylase UL114 with the viral DNA polymerase catalytic subunit UL54.J Gen Virol. 2010 Aug;91(Pt 8):2029-2033. doi: 10.1099/vir.0.022160-0. Epub 2010 Apr 21. J Gen Virol. 2010. PMID: 20410316 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

