Proteins that switch folds
- PMID: 20591649
- PMCID: PMC2928869
- DOI: 10.1016/j.sbi.2010.06.002
Proteins that switch folds
Abstract
An increasing number of proteins demonstrate the ability to switch between very different fold topologies, expanding their functional utility through new binding interactions. Recent examples of fold switching from naturally occurring and designed systems have a number of common features: (i) The structural transitions require states with diminished stability; (ii) Switching involves flexible regions in one conformer or the other; (iii) A new binding surface is revealed in the alternate fold that can lead to both stabilization of the alternative state and expansion of biological function. Fold switching not only provides insight into how new folds evolve, but also indicates that an amino acid sequence has more information content than previously thought. A polypeptide chain can encode a stable fold while simultaneously hiding latent propensities for alternative states with novel functions.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures
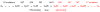
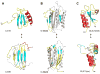
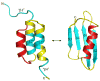
Similar articles
-
Reversible switching between two common protein folds in a designed system using only temperature.Proc Natl Acad Sci U S A. 2023 Jan 24;120(4):e2215418120. doi: 10.1073/pnas.2215418120. Epub 2023 Jan 20. Proc Natl Acad Sci U S A. 2023. PMID: 36669114 Free PMC article.
-
Local versus global fold switching in protein evolution: insight from a three-letter continuous model.Phys Biol. 2015 Feb 23;12(2):026002. doi: 10.1088/1478-3975/12/2/026002. Phys Biol. 2015. PMID: 25706822
-
Sequence and structural analysis of two designed proteins with 88% identity adopting different folds.Protein Eng Des Sel. 2010 Dec;23(12):911-8. doi: 10.1093/protein/gzq070. Epub 2010 Oct 15. Protein Eng Des Sel. 2010. PMID: 20952437
-
How does a knotted protein fold?FEBS J. 2009 Jan;276(2):365-75. doi: 10.1111/j.1742-4658.2008.06801.x. Epub 2008 Dec 10. FEBS J. 2009. PMID: 19077162 Review.
-
Inter-residue interactions in protein folding and stability.Prog Biophys Mol Biol. 2004 Oct;86(2):235-77. doi: 10.1016/j.pbiomolbio.2003.09.003. Prog Biophys Mol Biol. 2004. PMID: 15288760 Review.
Cited by
-
Intrinsic Disorder and Other Malleable Arsenals of Evolved Protein Multifunctionality.J Mol Evol. 2024 Dec;92(6):669-684. doi: 10.1007/s00239-024-10196-7. Epub 2024 Aug 30. J Mol Evol. 2024. PMID: 39214891 Review.
-
Information theoretic measures for quantifying sequence-ensemble relationships of intrinsically disordered proteins.Protein Eng Des Sel. 2019 Dec 31;32(4):191-202. doi: 10.1093/protein/gzz014. Protein Eng Des Sel. 2019. PMID: 31375817 Free PMC article.
-
Engineering protein assemblies with allosteric control via monomer fold-switching.Nat Commun. 2019 Dec 13;10(1):5703. doi: 10.1038/s41467-019-13686-1. Nat Commun. 2019. PMID: 31836707 Free PMC article.
-
Simulations of a protein fold switch reveal crowding-induced population shifts driven by disordered regions.Commun Chem. 2023 Sep 9;6(1):191. doi: 10.1038/s42004-023-00995-2. Commun Chem. 2023. PMID: 37689829 Free PMC article.
-
Common structural traits for cystine knot domain of the TGFβ superfamily of proteins and three-fingered ectodomain of their cellular receptors.Cell Mol Life Sci. 2011 Oct;68(20):3437-51. doi: 10.1007/s00018-011-0643-4. Epub 2011 Mar 3. Cell Mol Life Sci. 2011. PMID: 21369710 Free PMC article.
References
-
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. - PubMed
-
- Mottonen J, Strand A, Symersky J, Sweet RM, Danley DE, Geoghegan KF, Gerard RD, Goldsmith EJ. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355:270–273. - PubMed
-
- Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. - PubMed
-
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

