Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies
- PMID: 20584987
- PMCID: PMC2937563
- DOI: 10.1128/MCB.00429-10
Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies
Abstract
In eukaryotic cells, degradation of many mRNAs is initiated by removal of the poly(A) tail followed by decapping and 5'-3' exonucleolytic decay. Although the order of these events is well established, we are still lacking a mechanistic understanding of how deadenylation and decapping are linked. In this report we identify human Pat1b as a protein that is tightly associated with the Ccr4-Caf1-Not deadenylation complex as well as with the Dcp1-Dcp2 decapping complex. In addition, the RNA helicase Rck and Lsm1 proteins interact with human Pat1b. These interactions are mediated via at least three independent domains within Pat1b, suggesting that Pat1b serves as a scaffold protein. By tethering Pat1b to a reporter mRNA, we further provide evidence that Pat1b is also functionally linked to both deadenylation and decapping. Finally, we report that Pat1b strongly induces the formation of processing (P) bodies, cytoplasmic foci that contain most enzymes of the RNA decay machinery. An amino-terminal region within Pat1b serves as an aggregation-prone domain that nucleates P bodies, whereas an acidic domain controls the size of P bodies. Taken together, these findings provide evidence that human Pat1b is a central component of the RNA decay machinery by physically connecting deadenylation with decapping.
Figures

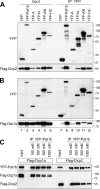
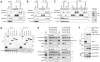
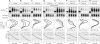
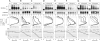
Similar articles
-
Role of Rck-Pat1b binding in assembly of processing-bodies.RNA Biol. 2013 Apr;10(4):528-39. doi: 10.4161/rna.24086. Epub 2013 Mar 27. RNA Biol. 2013. PMID: 23535175 Free PMC article.
-
A low-complexity region in human XRN1 directly recruits deadenylation and decapping factors in 5'-3' messenger RNA decay.Nucleic Acids Res. 2019 Sep 26;47(17):9282-9295. doi: 10.1093/nar/gkz633. Nucleic Acids Res. 2019. PMID: 31340047 Free PMC article.
-
HPat provides a link between deadenylation and decapping in metazoa.J Cell Biol. 2010 Apr 19;189(2):289-302. doi: 10.1083/jcb.200910141. J Cell Biol. 2010. PMID: 20404111 Free PMC article.
-
Pat1 RNA-binding proteins: Multitasking shuttling proteins.Wiley Interdiscip Rev RNA. 2019 Nov;10(6):e1557. doi: 10.1002/wrna.1557. Epub 2019 Jun 24. Wiley Interdiscip Rev RNA. 2019. PMID: 31231973 Review.
-
RNA decapping inside and outside of processing bodies.Curr Opin Cell Biol. 2005 Jun;17(3):326-31. doi: 10.1016/j.ceb.2005.04.002. Curr Opin Cell Biol. 2005. PMID: 15901504 Review.
Cited by
-
Structural and functional control of the eukaryotic mRNA decapping machinery.Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):580-9. doi: 10.1016/j.bbagrm.2012.12.006. Epub 2012 Dec 31. Biochim Biophys Acta. 2013. PMID: 23287066 Free PMC article. Review.
-
Artificial selection reveals the role of transcriptional constraints in the maintenance of life history variation.Evol Lett. 2020 Apr 7;4(3):200-211. doi: 10.1002/evl3.166. eCollection 2020 Jun. Evol Lett. 2020. PMID: 32547781 Free PMC article.
-
HELZ directly interacts with CCR4-NOT and causes decay of bound mRNAs.Life Sci Alliance. 2019 Sep 30;2(5):e201900405. doi: 10.26508/lsa.201900405. Print 2019 Oct. Life Sci Alliance. 2019. PMID: 31570513 Free PMC article.
-
Human 4E-T represses translation of bound mRNAs and enhances microRNA-mediated silencing.Nucleic Acids Res. 2014 Mar;42(5):3298-313. doi: 10.1093/nar/gkt1265. Epub 2013 Dec 13. Nucleic Acids Res. 2014. PMID: 24335285 Free PMC article.
-
Use of Cellular Decapping Activators by Positive-Strand RNA Viruses.Viruses. 2016 Dec 21;8(12):340. doi: 10.3390/v8120340. Viruses. 2016. PMID: 28009841 Free PMC article. Review.
References
-
- Bhattacharyya, S. N., R. Habermacher, U. Martine, E. I. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111-1124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
