Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design
- PMID: 20577269
- PMCID: PMC3167078
- DOI: 10.1038/nri2801
Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design
Abstract
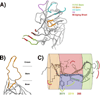
One of the main challenges of developing an HIV-1 vaccine lies in eliciting immune responses that can overcome the antigenic variability exhibited by HIV. Most HIV-1 vaccine development has focused on inducing immunity to conserved regions of the HIV-1 envelope. However, new studies of the sequence-variable regions of the HIV-1 gp120 envelope glycoprotein have shown that there are conserved immunological and structural features in these regions. Recombinant immunogens that include these features may provide the means to address the antigenic diversity of HIV-1 and induce protective antibodies that can prevent infection with HIV-1.
Figures


Similar articles
-
Rationally Designed Immunogens Targeting HIV-1 gp120 V1V2 Induce Distinct Conformation-Specific Antibody Responses in Rabbits.J Virol. 2016 Nov 28;90(24):11007-11019. doi: 10.1128/JVI.01409-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27707920 Free PMC article.
-
Rationally Designed Vaccines Targeting the V2 Region of HIV-1 gp120 Induce a Focused, Cross-Clade-Reactive, Biologically Functional Antibody Response.J Virol. 2016 Nov 28;90(24):10993-11006. doi: 10.1128/JVI.01403-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27630234 Free PMC article.
-
A Trimeric HIV-1 Envelope gp120 Immunogen Induces Potent and Broad Anti-V1V2 Loop Antibodies against HIV-1 in Rabbits and Rhesus Macaques.J Virol. 2018 Feb 12;92(5):e01796-17. doi: 10.1128/JVI.01796-17. Print 2018 Mar 1. J Virol. 2018. PMID: 29237847 Free PMC article.
-
GP120: target for neutralizing HIV-1 antibodies.Annu Rev Immunol. 2006;24:739-69. doi: 10.1146/annurev.immunol.24.021605.090557. Annu Rev Immunol. 2006. PMID: 16551265 Review.
-
[VLP vaccines and effects of HIV-1 Env protein modifications on their antigenic properties].Mol Biol (Mosk). 2016 May-Jun;50(3):406-15. doi: 10.7868/S0026898416030113. Mol Biol (Mosk). 2016. PMID: 27414779 Review. Russian.
Cited by
-
Glycan Masking of Epitopes in the NTD and RBD of the Spike Protein Elicits Broadly Neutralizing Antibodies Against SARS-CoV-2 Variants.Front Immunol. 2021 Dec 2;12:795741. doi: 10.3389/fimmu.2021.795741. eCollection 2021. Front Immunol. 2021. PMID: 34925381 Free PMC article.
-
Humoral Response to the HIV-1 Envelope V2 Region in a Thai Early Acute Infection Cohort.Cells. 2019 Apr 19;8(4):365. doi: 10.3390/cells8040365. Cells. 2019. PMID: 31010245 Free PMC article.
-
Editorial neuroAIDS review.AIDS. 2011 Jan 14;25(2):123-41. doi: 10.1097/QAD.0b013e328340fd42. AIDS. 2011. PMID: 21076277 Free PMC article. Review. No abstract available.
-
A conserved determinant in the V1 loop of HIV-1 modulates the V3 loop to prime low CD4 use and macrophage infection.J Virol. 2011 Mar;85(5):2397-405. doi: 10.1128/JVI.02187-10. Epub 2010 Dec 15. J Virol. 2011. PMID: 21159865 Free PMC article.
-
Structure of a cleavage-independent HIV Env recapitulates the glycoprotein architecture of the native cleaved trimer.Nat Commun. 2018 May 16;9(1):1956. doi: 10.1038/s41467-018-04272-y. Nat Commun. 2018. PMID: 29769533 Free PMC article.
References
-
- Gilbert PB, et al. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis. 2005;191:666–677. - PubMed
-
- Rerks-Ngarm S, et al. Vaccination with ALVAC and AIDSVAX to Prevent HIV-1 Infection in Thailand. N Engl J Med. 2009;361:2209–2220. - PubMed
-
- Primer V, editor. IAVI Report. 2010. Understanding Antibody FUnctions: Beyond Neutralization.
-
- Starcich BR, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI036085/AI/NIAID NIH HHS/United States
- R01 HL059725-03/HL/NHLBI NIH HHS/United States
- AI084119/AI/NIAID NIH HHS/United States
- R01 AI084119/AI/NIAID NIH HHS/United States
- DP2 OD004631-01/OD/NIH HHS/United States
- R01 AI084119-02/AI/NIAID NIH HHS/United States
- HL59725/HL/NHLBI NIH HHS/United States
- R01 HL059725-04/HL/NHLBI NIH HHS/United States
- DP2 OD004631/OD/NIH HHS/United States
- R01 AI084119-01/AI/NIAID NIH HHS/United States
- AI38065/AI/NIAID NIH HHS/United States
- R01 HL059725/HL/NHLBI NIH HHS/United States
- R01 AI036085-06/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

