WASP family proteins: their evolution and its physiological implications
- PMID: 20573979
- PMCID: PMC2921111
- DOI: 10.1091/mbc.E10-04-0372
WASP family proteins: their evolution and its physiological implications
Abstract
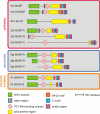
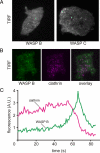
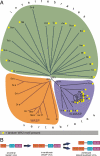
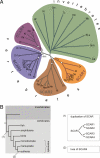
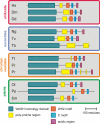
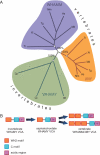
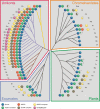
WASP family proteins control actin polymerization by activating the Arp2/3 complex. Several subfamilies exist, but their regulation and physiological roles are not well understood, nor is it even known if all subfamilies have been identified. Our extensive search reveals few novel WASP family proteins. The WASP, WASH, and SCAR/WAVE subfamilies are evolutionarily ancient, with WASH the most universally present, whereas WHAMM/JMY first appears in invertebrates. An unusual Dictyostelium WASP homologue that has lost the WH1 domain has retained its function in clathrin-mediated endocytosis, demonstrating that WASPs can function with a remarkably diverse domain topology. The WASH and SCAR/WAVE regulatory complexes are much more rigidly maintained; their domain topology is highly conserved, and all subunits are present or lost together, showing that the complexes are ancient and functionally interdependent. Finally, each subfamily has a distinctive C motif, indicating that this motif plays a specific role in each subfamily's function, unlike the generic V and A motifs. Our analysis identifies which features are universally conserved, and thus essential, and which are branch-specific modifications. It also shows the WASP family is more widespread and diverse than currently appreciated and unexpectedly biases the physiological role of the Arp2/3 complex toward vesicle traffic.
Figures




Similar articles
-
WASP family proteins: Molecular mechanisms and implications in human disease.Eur J Cell Biol. 2022 Jun-Aug;101(3):151244. doi: 10.1016/j.ejcb.2022.151244. Epub 2022 Jun 1. Eur J Cell Biol. 2022. PMID: 35667337 Free PMC article.
-
Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex.J Biol Chem. 2006 Apr 14;281(15):10589-97. doi: 10.1074/jbc.M507470200. Epub 2005 Dec 23. J Biol Chem. 2006. PMID: 16403731 Free PMC article.
-
Scar/WAVE drives actin protrusions independently of its VCA domain using proline-rich domains.Curr Biol. 2024 Oct 7;34(19):4436-4451.e9. doi: 10.1016/j.cub.2024.08.013. Epub 2024 Sep 26. Curr Biol. 2024. PMID: 39332399
-
New insights into the regulation and cellular functions of the ARP2/3 complex.Nat Rev Mol Cell Biol. 2013 Jan;14(1):7-12. doi: 10.1038/nrm3492. Epub 2012 Dec 5. Nat Rev Mol Cell Biol. 2013. PMID: 23212475 Review.
-
WASP family proteins, more than Arp2/3 activators.Biochem Soc Trans. 2016 Oct 15;44(5):1339-1345. doi: 10.1042/BST20160176. Biochem Soc Trans. 2016. PMID: 27911716 Free PMC article. Review.
Cited by
-
AlignNemo: a local network alignment method to integrate homology and topology.PLoS One. 2012;7(6):e38107. doi: 10.1371/journal.pone.0038107. Epub 2012 Jun 12. PLoS One. 2012. PMID: 22719866 Free PMC article.
-
The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIα.Mol Biol Cell. 2013 Jul;24(14):2269-84. doi: 10.1091/mbc.E13-02-0088. Epub 2013 May 15. Mol Biol Cell. 2013. PMID: 23676666 Free PMC article.
-
Differential Protein Profiling of Cerebrospinal Fluid in Piglets with Severe Meningoencephalitis Caused by Streptococcus suis Type 2 Compared to Controls.Front Cell Infect Microbiol. 2018 Feb 9;8:35. doi: 10.3389/fcimb.2018.00035. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29479521 Free PMC article.
-
A conformational change within the WAVE2 complex regulates its degradation following cellular activation.Sci Rep. 2017 Mar 23;7:44863. doi: 10.1038/srep44863. Sci Rep. 2017. PMID: 28332566 Free PMC article.
-
Sensitized mutagenesis screen in Factor V Leiden mice identifies thrombosis suppressor loci.Proc Natl Acad Sci U S A. 2017 Sep 5;114(36):9659-9664. doi: 10.1073/pnas.1705762114. Epub 2017 Aug 21. Proc Natl Acad Sci U S A. 2017. PMID: 28827327 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

