De novo characterization of a whitefly transcriptome and analysis of its gene expression during development
- PMID: 20573269
- PMCID: PMC2898760
- DOI: 10.1186/1471-2164-11-400
De novo characterization of a whitefly transcriptome and analysis of its gene expression during development
Abstract
Background: Whitefly (Bemisia tabaci) causes extensive crop damage throughout the world by feeding directly on plants and by vectoring hundreds of species of begomoviruses. Yet little is understood about its genes involved in development, insecticide resistance, host range plasticity and virus transmission.
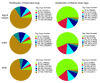
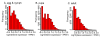
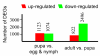
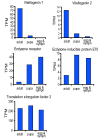
Results: To facilitate research on whitefly, we present a method for de novo assembly of whitefly transcriptome using short read sequencing technology (Illumina). In a single run, we produced more than 43 million sequencing reads. These reads were assembled into 168,900 unique sequences (mean size = 266 bp) which represent more than 10-fold of all the whitefly sequences deposited in the GenBank (as of March 2010). Based on similarity search with known proteins, these analyses identified 27,290 sequences with a cut-off E-value above 10-5. Assembled sequences were annotated with gene descriptions, gene ontology and clusters of orthologous group terms. In addition, we investigated the transcriptome changes during whitefly development using a tag-based digital gene expression (DGE) system. We obtained a sequencing depth of over 2.5 million tags per sample and identified a large number of genes associated with specific developmental stages and insecticide resistance.
Conclusion: Our data provides the most comprehensive sequence resource available for whitefly study and demonstrates that the Illumina sequencing allows de novo transcriptome assembly and gene expression analysis in a species lacking genome information. We anticipate that next generation sequencing technologies hold great potential for the study of the transcriptome in other non-model organisms.
Figures
Similar articles
-
Pyrosequencing the Bemisia tabaci transcriptome reveals a highly diverse bacterial community and a robust system for insecticide resistance.PLoS One. 2012;7(4):e35181. doi: 10.1371/journal.pone.0035181. Epub 2012 Apr 30. PLoS One. 2012. PMID: 22558125 Free PMC article.
-
Transcriptome and gene expression analysis of the rice leaf folder, Cnaphalocrosis medinalis.PLoS One. 2012;7(11):e47401. doi: 10.1371/journal.pone.0047401. Epub 2012 Nov 19. PLoS One. 2012. PMID: 23185238 Free PMC article.
-
Analysis of a native whitefly transcriptome and its sequence divergence with two invasive whitefly species.BMC Genomics. 2012 Oct 4;13:529. doi: 10.1186/1471-2164-13-529. BMC Genomics. 2012. PMID: 23036081 Free PMC article.
-
Transcriptome analysis and comparison reveal divergence between two invasive whitefly cryptic species.BMC Genomics. 2011 Sep 22;12:458. doi: 10.1186/1471-2164-12-458. BMC Genomics. 2011. PMID: 21939539 Free PMC article.
-
Host plant adaptation in the polyphagous whitefly, Trialeurodes vaporariorum, is associated with transcriptional plasticity and altered sensitivity to insecticides.BMC Genomics. 2019 Dec 19;20(1):996. doi: 10.1186/s12864-019-6397-3. BMC Genomics. 2019. PMID: 31856729 Free PMC article.
Cited by
-
Comparative analysis of differentially expressed genes in Sika deer antler at different stages.Mol Biol Rep. 2013 Feb;40(2):1665-76. doi: 10.1007/s11033-012-2216-5. Epub 2012 Oct 18. Mol Biol Rep. 2013. PMID: 23073784
-
Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study.BMC Bioinformatics. 2011 Dec 14;12 Suppl 14(Suppl 14):S2. doi: 10.1186/1471-2105-12-S14-S2. BMC Bioinformatics. 2011. PMID: 22373417 Free PMC article.
-
Analysis of the transcriptional differences between indigenous and invasive whiteflies reveals possible mechanisms of whitefly invasion.PLoS One. 2013 May 8;8(5):e62176. doi: 10.1371/journal.pone.0062176. Print 2013. PLoS One. 2013. PMID: 23667457 Free PMC article.
-
Unraveling dual feeding associated molecular complexity of salivary glands in the mosquito Anopheles culicifacies.Biol Open. 2015 Jul 10;4(8):1002-15. doi: 10.1242/bio.012294. Biol Open. 2015. PMID: 26163527 Free PMC article.
-
Comparative transcript profiling of the fertile and sterile flower buds of pol CMS in B. napus.BMC Genomics. 2014 Apr 3;15:258. doi: 10.1186/1471-2164-15-258. BMC Genomics. 2014. PMID: 24707970 Free PMC article.
References
-
- Boykin LM, Shatters RG, Rosell RC, McKenzie CL, Bagnall RA, De Barro PJ, Frohlich DR. Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol Phylogenet Evol. 2007;44:1306–1319. doi: 10.1016/j.ympev.2007.04.020. - DOI - PubMed
-
- Czosnek H, Ghanim M, Ghanim M. The circulative pathway of begomoviruses in the whitefly vector Bemisia tabaci; insights from studies with Tomato yellow leaf curl virus. Ann Appl Biol. 2002;140:215–231. doi: 10.1111/j.1744-7348.2002.tb00175.x. - DOI
-
- Seal S, vandenBosch F, Jeger M. Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit Rev Plant Sci. 2006;25:23–46. doi: 10.1080/07352680500365257. - DOI
-
- Jiu M, Zhou XP, Liu SS. Acquisition and transmission of two begomoviruses by the B and a non-B biotype of Bemisia tabaci from Zhejiang, China. Journal of Phytopathology. 2006;154:587–591. doi: 10.1111/j.1439-0434.2006.01151.x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

