Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells
- PMID: 20554777
- PMCID: PMC2919005
- DOI: 10.1128/JVI.00456-10
Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells
Abstract
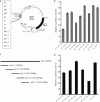
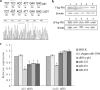
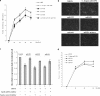
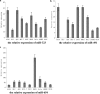
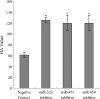
MicroRNAs (miRNAs) are a class of noncoding RNAs of lengths ranging from 18 to 23 nucleotides (nt) that play critical roles in a wide variety of biological processes. There is a growing amount of evidence that miRNAs play critical roles in intricate host-pathogen interaction networks, but the involvement of miRNAs during influenza viral infection is unknown. To determine whether the cellular miRNAs play an important role in H1N1 influenza A viral infections, 3' untranslated region (UTR) reporter analysis was used to identify putative miRNA targets in the influenza virus genome, and virus proliferation analysis was used to detect the effect of the screened miRNAs on the replication of H1N1 influenza A virus (A/WSN/33) in MDCK cells. The results showed that miRNA 323 (miR-323), miR-491, and miR-654 inhibit replication of the H1N1 influenza A virus through binding to the PB1 gene. Moreover mutational analysis of the predicted miRNA binding sites showed that the three miRNAs bind to the same conserved region of the PB1 gene. Intriguingly, despite the fact that the miRNAs and PB1 mRNA binding sequences are not a perfect match, the miRNAs downregulate PB1 expression through mRNA degradation instead of translation repression. This is the first demonstration that cellular miRNAs regulate influenza viral replication by degradation of the viral gene. Our findings support the notion that any miRNA has antiviral potential, independent of its cellular function, and that the cellular miRNAs play an important role in the host, defending against virus infection.
Figures


Similar articles
-
Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells.J Cell Mol Med. 2012 Oct;16(10):2539-46. doi: 10.1111/j.1582-4934.2012.01572.x. J Cell Mol Med. 2012. PMID: 22452878 Free PMC article.
-
MicroRNA hsa-miR-324-5p Suppresses H5N1 Virus Replication by Targeting the Viral PB1 and Host CUEDC2.J Virol. 2018 Sep 12;92(19):e01057-18. doi: 10.1128/JVI.01057-18. Print 2018 Oct 1. J Virol. 2018. PMID: 30045983 Free PMC article.
-
The highly pathogenic H5N1 influenza A virus down-regulated several cellular MicroRNAs which target viral genome.J Cell Mol Med. 2017 Nov;21(11):3076-3086. doi: 10.1111/jcmm.13219. Epub 2017 Jun 13. J Cell Mol Med. 2017. PMID: 28609011 Free PMC article.
-
Critical role of microRNAs in host and influenza A (H1N1) virus interactions.Life Sci. 2021 Jul 15;277:119484. doi: 10.1016/j.lfs.2021.119484. Epub 2021 Apr 13. Life Sci. 2021. PMID: 33862119 Review.
-
Host microRNAs and exosomes that modulate influenza virus infection.Virus Res. 2020 Apr 2;279:197885. doi: 10.1016/j.virusres.2020.197885. Epub 2020 Jan 22. Virus Res. 2020. PMID: 31981772 Review.
Cited by
-
Cellular microRNA miR-26a suppresses replication of porcine reproductive and respiratory syndrome virus by activating innate antiviral immunity.Sci Rep. 2015 May 27;5:10651. doi: 10.1038/srep10651. Sci Rep. 2015. PMID: 26013676 Free PMC article.
-
Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients.Transl Res. 2021 Oct;236:147-159. doi: 10.1016/j.trsl.2021.05.004. Epub 2021 May 26. Transl Res. 2021. PMID: 34048985 Free PMC article. Clinical Trial.
-
The role of microRNAs in COVID-19 with a focus on miR-200c.J Circ Biomark. 2022 Mar 21;11:14-23. doi: 10.33393/jcb.2022.2356. eCollection 2022 Jan-Dec. J Circ Biomark. 2022. PMID: 35356072 Free PMC article. Review.
-
On the Importance of Host MicroRNAs During Viral Infection.Front Genet. 2018 Oct 2;9:439. doi: 10.3389/fgene.2018.00439. eCollection 2018. Front Genet. 2018. PMID: 30333857 Free PMC article. Review.
-
Cellular microRNA miR-181b inhibits replication of mink enteritis virus by repression of non-structural protein 1 translation.PLoS One. 2013 Dec 11;8(12):e81515. doi: 10.1371/journal.pone.0081515. eCollection 2013. PLoS One. 2013. PMID: 24349084 Free PMC article.
References
-
- Bartel, D. P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281-297. - PubMed
-
- Bennasser, Y., S. Y. Le, M. Benkirane, and K. T. Jeang. 2005. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22:607-619. - PubMed
-
- Bentwich, I. 2005. Prediction and validation of microRNAs and their targets. FEBS Lett. 579:5904-5910. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

