Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP
- PMID: 20548956
- PMCID: PMC2883600
- DOI: 10.1371/journal.ppat.1000951
Epigenetic repression of p16(INK4A) by latent Epstein-Barr virus requires the interaction of EBNA3A and EBNA3C with CtBP
Abstract
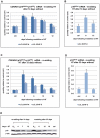
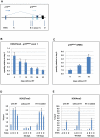
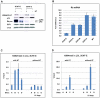
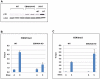
As an inhibitor of cyclin-dependent kinases, p16(INK4A) is an important tumour suppressor and inducer of cellular senescence that is often inactivated during the development of cancer by promoter DNA methylation. Using newly established lymphoblastoid cell lines (LCLs) expressing a conditional EBNA3C from recombinant EBV, we demonstrate that EBNA3C inactivation initiates chromatin remodelling that resets the epigenetic status of p16(INK4A) to permit transcriptional activation: the polycomb-associated repressive H3K27me3 histone modification is substantially reduced, while the activation-related mark H3K4me3 is modestly increased. Activation of EBNA3C reverses the distribution of these epigenetic marks, represses p16(INK4A) transcription and allows proliferation. LCLs lacking EBNA3A express relatively high levels of p16(INK4A) and have a similar pattern of histone modifications on p16(INK4A) as produced by the inactivation of EBNA3C. Since binding to the co-repressor of transcription CtBP has been linked to the oncogenic activity of EBNA3A and EBNA3C, we established LCLs with recombinant viruses encoding EBNA3A- and/or EBNA3C-mutants that no longer bind CtBP. These novel LCLs have revealed that the chromatin remodelling and epigenetic repression of p16(INK4A) requires the interaction of both EBNA3A and EBNA3C with CtBP. The repression of p16(INK4A) by latent EBV will not only overcome senescence in infected B cells, but may also pave the way for p16(INK4A) DNA methylation during B cell lymphomagenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Induction of p16(INK4a) is the major barrier to proliferation when Epstein-Barr virus (EBV) transforms primary B cells into lymphoblastoid cell lines.PLoS Pathog. 2013 Feb;9(2):e1003187. doi: 10.1371/journal.ppat.1003187. Epub 2013 Feb 21. PLoS Pathog. 2013. PMID: 23436997 Free PMC article.
-
Epstein-Barr Virus (EBV) Latent Protein EBNA3A Directly Targets and Silences the STK39 Gene in B Cells Infected by EBV.J Virol. 2018 Mar 14;92(7):e01918-17. doi: 10.1128/JVI.01918-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29367247 Free PMC article.
-
Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression.Proc Natl Acad Sci U S A. 2011 Feb 1;108(5):1919-24. doi: 10.1073/pnas.1019599108. Epub 2011 Jan 18. Proc Natl Acad Sci U S A. 2011. PMID: 21245331 Free PMC article.
-
The EBNA3 Family: Two Oncoproteins and a Tumour Suppressor that Are Central to the Biology of EBV in B Cells.Curr Top Microbiol Immunol. 2015;391:61-117. doi: 10.1007/978-3-319-22834-1_3. Curr Top Microbiol Immunol. 2015. PMID: 26428372 Review.
-
Nucleoside diphosphate kinase/Nm23 and Epstein-Barr virus.Mol Cell Biochem. 2009 Sep;329(1-2):131-9. doi: 10.1007/s11010-009-0123-4. Epub 2009 May 3. Mol Cell Biochem. 2009. PMID: 19412732 Free PMC article. Review.
Cited by
-
Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation.J Virol. 2012 Oct;86(20):11096-106. doi: 10.1128/JVI.01069-12. Epub 2012 Aug 1. J Virol. 2012. PMID: 22855490 Free PMC article.
-
Epstein-Barr virus nuclear antigen 3C binds to BATF/IRF4 or SPI1/IRF4 composite sites and recruits Sin3A to repress CDKN2A.Proc Natl Acad Sci U S A. 2014 Jan 7;111(1):421-6. doi: 10.1073/pnas.1321704111. Epub 2013 Dec 16. Proc Natl Acad Sci U S A. 2014. PMID: 24344258 Free PMC article.
-
Viral-Targeted Strategies Against EBV-Associated Lymphoproliferative Diseases.Front Oncol. 2019 Feb 26;9:81. doi: 10.3389/fonc.2019.00081. eCollection 2019. Front Oncol. 2019. PMID: 30873380 Free PMC article. Review.
-
EBV finds a polycomb-mediated, epigenetic solution to the problem of oncogenic stress responses triggered by infection.Front Genet. 2013 Oct 24;4:212. doi: 10.3389/fgene.2013.00212. Front Genet. 2013. PMID: 24167519 Free PMC article. Review.
-
An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells.Cell Host Microbe. 2010 Dec 16;8(6):510-22. doi: 10.1016/j.chom.2010.11.004. Cell Host Microbe. 2010. PMID: 21147465 Free PMC article.
References
-
- Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr Virus and the Origins of Associated Lymphomas. The New England journal of medicine. 2004;350:1328–1337. - PubMed
-
- Touitou R, O'Nions J, Heaney J, Allday MJ. Epstein-Barr virus EBNA3 proteins bind to the C8/alpha7 subunit of the 20S proteasome and are degraded by 20S proteasomes in vitro, but are very stable in latently infected B cells. J Gen Virol. 2005;86:1269–1277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

