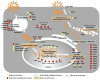
Cellular networks involved in the influenza virus life cycle
- PMID: 20542247
- PMCID: PMC3167038
- DOI: 10.1016/j.chom.2010.05.008
Cellular networks involved in the influenza virus life cycle
Abstract
Influenza viruses cause epidemics and pandemics. Like all viruses, influenza viruses rely on the host cellular machinery to support their life cycle. Accordingly, identification of the host functions co-opted for viral replication is of interest to understand the mechanisms of the virus life cycle and to find new targets for the development of antiviral compounds. Among the various approaches used to explore host factor involvement in the influenza virus replication cycle, perhaps the most powerful is RNAi-based genome-wide screening, which has shed new light on the search for host factors involved in virus replication. In this review, we examine the cellular genes identified to date as important for influenza virus replication in genome-wide screens, assess pathways that were repeatedly identified in these studies, and discuss how these pathways might be involved in the individual steps of influenza virus replication, ultimately leading to a comprehensive understanding of the virus life cycle.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Influenza virus-host interactomes as a basis for antiviral drug development.Curr Opin Virol. 2015 Oct;14:71-8. doi: 10.1016/j.coviro.2015.08.008. Epub 2015 Sep 13. Curr Opin Virol. 2015. PMID: 26364134 Free PMC article. Review.
-
Role of Host Genes in Influenza Virus Replication.Curr Top Microbiol Immunol. 2018;419:151-189. doi: 10.1007/82_2017_30. Curr Top Microbiol Immunol. 2018. PMID: 28643205 Review.
-
Network-Guided Discovery of Influenza Virus Replication Host Factors.mBio. 2018 Dec 18;9(6):e02002-18. doi: 10.1128/mBio.02002-18. mBio. 2018. PMID: 30563907 Free PMC article.
-
Uncovering the global host cell requirements for influenza virus replication via RNAi screening.Microbes Infect. 2011 May;13(5):516-25. doi: 10.1016/j.micinf.2011.01.012. Epub 2011 Jan 27. Microbes Infect. 2011. PMID: 21276872 Free PMC article. Review.
-
Interplay between host non-coding RNAs and influenza viruses.RNA Biol. 2021 May;18(5):767-784. doi: 10.1080/15476286.2021.1872170. Epub 2021 Jan 18. RNA Biol. 2021. PMID: 33404285 Free PMC article. Review.
Cited by
-
Host modulators of H1N1 cytopathogenicity.PLoS One. 2012;7(8):e39284. doi: 10.1371/journal.pone.0039284. Epub 2012 Aug 2. PLoS One. 2012. PMID: 22876275 Free PMC article.
-
Computational design of host transcription-factors sets whose misregulation mimics the transcriptomic effect of viral infections.Sci Rep. 2012;2:1006. doi: 10.1038/srep01006. Epub 2012 Dec 19. Sci Rep. 2012. PMID: 23256040 Free PMC article.
-
HIF-1α promotes virus replication and cytokine storm in H1N1 virus-induced severe pneumonia through cellular metabolic reprogramming.Virol Sin. 2024 Feb;39(1):81-96. doi: 10.1016/j.virs.2023.11.010. Epub 2023 Nov 30. Virol Sin. 2024. PMID: 38042371 Free PMC article.
-
Nasal symbiont Staphylococcus epidermidis restricts the cellular entry of influenza virus into the nasal epithelium.NPJ Biofilms Microbiomes. 2022 Apr 13;8(1):26. doi: 10.1038/s41522-022-00290-3. NPJ Biofilms Microbiomes. 2022. PMID: 35418111 Free PMC article.
-
Viral-host interactions during splicing and nuclear export of influenza virus mRNAs.Curr Opin Virol. 2022 Aug;55:101254. doi: 10.1016/j.coviro.2022.101254. Epub 2022 Jul 29. Curr Opin Virol. 2022. PMID: 35908311 Free PMC article. Review.
References
-
- Barman S, Ali A, Hui EK, Adhikary L, Nayak DP. Transport of viral proteins to the apical membranes and interaction of matrix protein with glycoproteins in the assembly of influenza viruses. Virus Res. 2001;77:61–69. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

