A survey of genomic traces reveals a common sequencing error, RNA editing, and DNA editing
- PMID: 20531933
- PMCID: PMC2873906
- DOI: 10.1371/journal.pgen.1000954
A survey of genomic traces reveals a common sequencing error, RNA editing, and DNA editing
Abstract
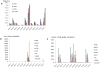
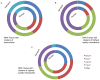
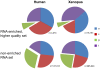
While it is widely held that an organism's genomic information should remain constant, several protein families are known to modify it. Members of the AID/APOBEC protein family can deaminate DNA. Similarly, members of the ADAR family can deaminate RNA. Characterizing the scope of these events is challenging. Here we use large genomic data sets, such as the two billion sequences in the NCBI Trace Archive, to look for clusters of mismatches of the same type, which are a hallmark of editing events caused by APOBEC3 and ADAR. We align 603,249,815 traces from the NCBI trace archive to their reference genomes. In clusters of mismatches of increasing size, at least one systematic sequencing error dominates the results (G-to-A). It is still present in mismatches with 99% accuracy and only vanishes in mismatches at 99.99% accuracy or higher. The error appears to have entered into about 1% of the HapMap, possibly affecting other users that rely on this resource. Further investigation, using stringent quality thresholds, uncovers thousands of mismatch clusters with no apparent defects in their chromatograms. These traces provide the first reported candidates of endogenous DNA editing in human, further elucidating RNA editing in human and mouse and also revealing, for the first time, extensive RNA editing in Xenopus tropicalis. We show that the NCBI Trace Archive provides a valuable resource for the investigation of the phenomena of DNA and RNA editing, as well as setting the stage for a comprehensive mapping of editing events in large-scale genomic datasets.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Prediction of constitutive A-to-I editing sites from human transcriptomes in the absence of genomic sequences.BMC Genomics. 2013 Mar 27;14:206. doi: 10.1186/1471-2164-14-206. BMC Genomics. 2013. PMID: 23537002 Free PMC article.
-
A cytosine deaminase for programmable single-base RNA editing.Science. 2019 Jul 26;365(6451):382-386. doi: 10.1126/science.aax7063. Epub 2019 Jul 11. Science. 2019. PMID: 31296651 Free PMC article.
-
Mutagenesis of apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, reveals distinct domains that mediate cytosine nucleoside deaminase, RNA binding, and RNA editing activity.J Biol Chem. 1995 Jun 16;270(24):14768-75. J Biol Chem. 1995. PMID: 7782343
-
The emerging role of RNA and DNA editing in cancer.Biochim Biophys Acta. 2014 Apr;1845(2):308-16. doi: 10.1016/j.bbcan.2014.03.001. Epub 2014 Mar 7. Biochim Biophys Acta. 2014. PMID: 24607277 Review.
-
Diverse functions for DNA and RNA editing in the immune system.RNA Biol. 2010 Mar-Apr;7(2):220-8. doi: 10.4161/rna.7.2.11344. Epub 2010 Mar 29. RNA Biol. 2010. PMID: 20220309 Review.
Cited by
-
Detection of murine leukemia virus in the Epstein-Barr virus-positive human B-cell line JY, using a computational RNA-Seq-based exogenous agent detection pipeline, PARSES.J Virol. 2012 Mar;86(6):2970-7. doi: 10.1128/JVI.06717-11. Epub 2012 Jan 11. J Virol. 2012. PMID: 22238296 Free PMC article.
-
An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals.BMC Evol Biol. 2012 May 28;12:71. doi: 10.1186/1471-2148-12-71. BMC Evol Biol. 2012. PMID: 22640020 Free PMC article.
-
Requirement of the RNA-editing enzyme ADAR2 for normal physiology in mice.J Biol Chem. 2011 May 27;286(21):18614-22. doi: 10.1074/jbc.M110.200881. Epub 2011 Apr 5. J Biol Chem. 2011. PMID: 21467037 Free PMC article.
-
Massive A-to-I RNA editing is common across the Metazoa and correlates with dsRNA abundance.Genome Biol. 2017 Oct 2;18(1):185. doi: 10.1186/s13059-017-1315-y. Genome Biol. 2017. PMID: 28969707 Free PMC article.
-
Evaluation of the Interplay between the ADAR Editome and Immunotherapy in Melanoma.Noncoding RNA. 2021 Jan 12;7(1):5. doi: 10.3390/ncrna7010005. Noncoding RNA. 2021. PMID: 33445472 Free PMC article.
References
-
- Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, et al. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

