Suppression of the novel ER protein Maxer by mutant ataxin-1 in Bergman glia contributes to non-cell-autonomous toxicity
- PMID: 20531390
- PMCID: PMC2910266
- DOI: 10.1038/emboj.2010.116
Suppression of the novel ER protein Maxer by mutant ataxin-1 in Bergman glia contributes to non-cell-autonomous toxicity
Abstract
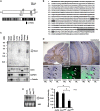
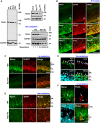
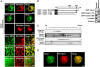
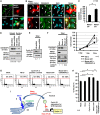
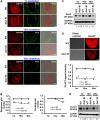
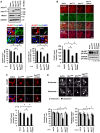
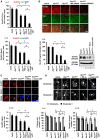
Non-cell-autonomous effect of mutant proteins expressed in glia has been implicated in several neurodegenerative disorders, whereas molecules mediating the toxicity are currently not known. We identified a novel molecule named multiple alpha-helix protein located at ER (Maxer) downregulated by mutant ataxin-1 (Atx1) in Bergmann glia. Maxer is an endoplasmic reticulum (ER) membrane protein interacting with CDK5RAP3. Maxer anchors CDK5RAP3 to the ER and inhibits its function of Cyclin D1 transcription repression in the nucleus. The loss of Maxer eventually induces cell accumulation at G1 phase. It was also shown that mutant Atx1 represses Maxer and inhibits proliferation of Bergmann glia in vitro. Consistently, Bergmann glia are reduced in the cerebellum of mutant Atx1 knockin mice before onset. Glutamate-aspartate transporter reduction in Bergmann glia by mutant Atx1 and vulnerability of Purkinje cell to glutamate are both strengthened by Maxer knockdown in Bergmann glia, whereas Maxer overexpression rescues them. Collectively, these results suggest that the reduction of Maxer mediates functional deficiency of Bergmann glia, and might contribute to the non-cell-autonomous pathology of SCA1.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Similar articles
-
Bergmann glia expression of polyglutamine-expanded ataxin-7 produces neurodegeneration by impairing glutamate transport.Nat Neurosci. 2006 Oct;9(10):1302-11. doi: 10.1038/nn1750. Epub 2006 Aug 27. Nat Neurosci. 2006. PMID: 16936724
-
Chronic optogenetic stimulation of Bergman glia leads to dysfunction of EAAT1 and Purkinje cell death, mimicking the events caused by expression of pathogenic ataxin-1.Neurobiol Dis. 2021 Jul;154:105340. doi: 10.1016/j.nbd.2021.105340. Epub 2021 Mar 19. Neurobiol Dis. 2021. PMID: 33753288
-
Decreased expression of glutamate transporter GLAST in Bergmann glia is associated with the loss of Purkinje neurons in the spinocerebellar ataxia type 1.Cerebellum. 2015 Feb;14(1):8-11. doi: 10.1007/s12311-014-0605-0. Cerebellum. 2015. PMID: 25255716
-
Progress in pathogenesis studies of spinocerebellar ataxia type 1.Philos Trans R Soc Lond B Biol Sci. 1999 Jun 29;354(1386):1079-81. doi: 10.1098/rstb.1999.0462. Philos Trans R Soc Lond B Biol Sci. 1999. PMID: 10434309 Free PMC article. Review.
-
Spinocerebellar ataxia type 1--modeling the pathogenesis of a polyglutamine neurodegenerative disorder in transgenic mice.J Neuropathol Exp Neurol. 2000 Apr;59(4):265-70. doi: 10.1093/jnen/59.4.265. J Neuropathol Exp Neurol. 2000. PMID: 10759181 Review.
Cited by
-
CDK5RAP3 is a novel repressor of p14ARF in hepatocellular carcinoma cells.PLoS One. 2012;7(7):e42210. doi: 10.1371/journal.pone.0042210. Epub 2012 Jul 31. PLoS One. 2012. PMID: 22860085 Free PMC article.
-
Brain region specific pre-synaptic and post-synaptic degeneration are early components of neuropathology in prion disease.PLoS One. 2013;8(1):e55004. doi: 10.1371/journal.pone.0055004. Epub 2013 Jan 30. PLoS One. 2013. PMID: 23383030 Free PMC article.
-
Developmental YAPdeltaC determines adult pathology in a model of spinocerebellar ataxia type 1.Nat Commun. 2017 Nov 30;8(1):1864. doi: 10.1038/s41467-017-01790-z. Nat Commun. 2017. PMID: 29192206 Free PMC article.
-
Biallelic Variants in UBA5 Reveal that Disruption of the UFM1 Cascade Can Result in Early-Onset Encephalopathy.Am J Hum Genet. 2016 Sep 1;99(3):695-703. doi: 10.1016/j.ajhg.2016.06.030. Epub 2016 Aug 18. Am J Hum Genet. 2016. PMID: 27545681 Free PMC article.
-
Viral Vector-Based Dissection of Marmoset GFAP Promoter in Mouse and Marmoset Brains.PLoS One. 2016 Aug 29;11(8):e0162023. doi: 10.1371/journal.pone.0162023. eCollection 2016. PLoS One. 2016. PMID: 27571575 Free PMC article.
References
-
- Alcock J, Scotting P, Sottile V (2007) Bergmann glia as putative stem cells of the mature cerebellum. Med Hypotheses 69: 341–345 - PubMed
-
- Basco E, Hajos F, Fulop Z (1977) Proliferation of Bergmann-glia in the developing rat cerebellum. Anat Embryol (Berl) 151: 219–222 - PubMed
-
- Cao Y, Bonizzi G, Seagroves TN, Greten FR, Johnson R, Schmidt EV, Karin M (2001) IKKalpha provides an essential link between RANK signaling and cyclin D1 expression during mammary gland development. Cell 107: 763–775 - PubMed
-
- Ching YP, Qi Z, Wang JH (2000) Cloning of three novel neuronal Cdk5 activator binding proteins. Gene 242: 285–294 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

