Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance
- PMID: 20519119
- PMCID: PMC2881480
- DOI: 10.1016/j.cmet.2010.04.005
Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance
Abstract
Autophagy is a homeostatic process involved in the bulk degradation of cytoplasmic components, including damaged organelles and proteins. In both genetic and dietary models of obesity, we observed a severe downregulation of autophagy, particularly in Atg7 expression levels in liver. Suppression of Atg7 both in vitro and in vivo resulted in defective insulin signaling and elevated ER stress. In contrast, restoration of the Atg7 expression in liver resulted in dampened ER stress, enhanced hepatic insulin action, and systemic glucose tolerance in obese mice. The beneficial action of Atg7 restoration in obese mice could be completely prevented by blocking a downstream mediator, Atg5, supporting its dependence on autophagy in regulating insulin action. Our data demonstrate that autophagy is an important regulator of organelle function and insulin signaling and that loss of autophagy is a critical component of defective insulin action seen in obesity.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures
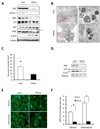
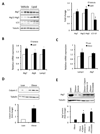

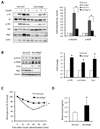

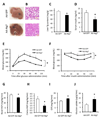
Comment in
-
Autophagy: a potential link between obesity and insulin resistance.Cell Metab. 2010 Jun 9;11(6):449-51. doi: 10.1016/j.cmet.2010.05.006. Cell Metab. 2010. PMID: 20519116
Similar articles
-
Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance.Mol Pharmacol. 2009 Sep;76(3):596-603. doi: 10.1124/mol.109.057067. Epub 2009 Jun 18. Mol Pharmacol. 2009. PMID: 19541767 Free PMC article.
-
Deficiency in the mitochondrial apoptotic pathway reveals the toxic potential of autophagy under ER stress conditions.Autophagy. 2014;10(11):1921-36. doi: 10.4161/15548627.2014.981790. Autophagy. 2014. PMID: 25470234 Free PMC article.
-
4-PBA reverses autophagic dysfunction and improves insulin sensitivity in adipose tissue of obese mice via Akt/mTOR signaling.Biochem Biophys Res Commun. 2017 Mar 11;484(3):529-535. doi: 10.1016/j.bbrc.2017.01.106. Epub 2017 Jan 30. Biochem Biophys Res Commun. 2017. PMID: 28153729
-
Endoplasmic reticulum proteostasis in hepatic steatosis.Nat Rev Endocrinol. 2016 Dec;12(12):710-722. doi: 10.1038/nrendo.2016.124. Epub 2016 Aug 12. Nat Rev Endocrinol. 2016. PMID: 27516341 Review.
-
Role of autophagy in diabetes and mitochondria.Ann N Y Acad Sci. 2010 Jul;1201:79-83. doi: 10.1111/j.1749-6632.2010.05614.x. Ann N Y Acad Sci. 2010. PMID: 20649543 Review.
Cited by
-
Autophagy, myocardial protection, and the metabolic syndrome.J Cardiovasc Pharmacol. 2012 Aug;60(2):125-32. doi: 10.1097/FJC.0b013e318256ce10. J Cardiovasc Pharmacol. 2012. PMID: 22472909 Free PMC article. Review.
-
Proteasome dysfunction mediates obesity-induced endoplasmic reticulum stress and insulin resistance in the liver.Diabetes. 2013 Mar;62(3):811-24. doi: 10.2337/db11-1652. Epub 2012 Dec 3. Diabetes. 2013. PMID: 23209186 Free PMC article.
-
Nutritional status and cardiac autophagy.Diabetes Metab J. 2013 Feb;37(1):30-5. doi: 10.4093/dmj.2013.37.1.30. Epub 2013 Feb 15. Diabetes Metab J. 2013. PMID: 23441078 Free PMC article.
-
Mapping autophagy on to your metabolic radar.Diabetes. 2012 Feb;61(2):272-80. doi: 10.2337/db11-1199. Diabetes. 2012. PMID: 22275084 Free PMC article. Review. No abstract available.
-
Sex-Specific Regulation of Interferon-γ Cytotoxicity in Mouse Liver by Autophagy.Hepatology. 2021 Nov;74(5):2745-2758. doi: 10.1002/hep.32010. Epub 2021 Aug 30. Hepatology. 2021. PMID: 34118081 Free PMC article.
References
-
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. - PubMed
-
- Ding WX, Yin XM. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy. 2008;4:141–150. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

