The miR-144/451 locus is required for erythroid homeostasis
- PMID: 20513743
- PMCID: PMC2901075
- DOI: 10.1084/jem.20100458
The miR-144/451 locus is required for erythroid homeostasis
Abstract
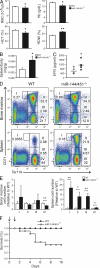
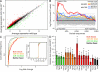
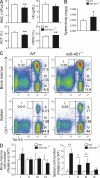
The process of erythropoiesis must be efficient and robust to supply the organism with red bloods cells both under condition of homeostasis and stress. The microRNA (miRNA) pathway was recently shown to regulate erythroid development. Here, we show that expression of the locus encoding miR-144 and miR-451 is strictly dependent on Argonaute 2 and is required for erythroid homeostasis. Mice deficient for the miR-144/451 cluster display a cell autonomous impairment of late erythroblast maturation, resulting in erythroid hyperplasia, splenomegaly, and a mild anemia. Analysis of gene expression profiles from wild-type and miR-144/451-deficient erythroblasts revealed that the miR-144/451 cluster acts as a "tuner" of gene expression, influencing the expression of many genes. MiR-451 imparts a greater impact on target gene expression than miR-144. Accordingly, mice deficient in miR-451 alone exhibited a phenotype indistinguishable from miR-144/451-deficient mice. Thus, the miR-144/451 cluster tunes gene expression to impart a robustness to erythropoiesis that is critical under conditions of stress.
Figures


Similar articles
-
Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta.Genes Dev. 2010 Aug 1;24(15):1614-9. doi: 10.1101/gad.1942810. Genes Dev. 2010. PMID: 20679397 Free PMC article.
-
Expression of microRNA-155 in thalassemic erythropoiesis.PeerJ. 2024 Sep 20;12:e18054. doi: 10.7717/peerj.18054. eCollection 2024. PeerJ. 2024. PMID: 39314840 Free PMC article.
-
miR-144/451 inhibits c-Myc to promote erythroid differentiation.FASEB J. 2020 Oct;34(10):13194-13210. doi: 10.1096/fj.202000941R. Epub 2020 Aug 16. FASEB J. 2020. PMID: 33319407
-
A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway.Genes Dev. 2007 Aug 15;21(16):1999-2004. doi: 10.1101/gad.1565607. Epub 2007 Jul 12. Genes Dev. 2007. PMID: 17626790 Free PMC article.
-
Involvement of miRNA in erythroid differentiation.Epigenomics. 2012 Feb;4(1):51-65. doi: 10.2217/epi.11.104. Epigenomics. 2012. PMID: 22332658 Review.
Cited by
-
Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations.Proc Natl Acad Sci U S A. 2013 Mar 12;110(11):4255-60. doi: 10.1073/pnas.1214046110. Epub 2013 Feb 25. Proc Natl Acad Sci U S A. 2013. PMID: 23440203 Free PMC article. Clinical Trial.
-
c-Myc suppresses miR-451⊣YWTAZ/AKT axis via recruiting HDAC3 in acute myeloid leukemia.Oncotarget. 2016 Nov 22;7(47):77430-77443. doi: 10.18632/oncotarget.12679. Oncotarget. 2016. PMID: 27764807 Free PMC article.
-
Evolution after Whole-Genome Duplication: Teleost MicroRNAs.Mol Biol Evol. 2021 Jul 29;38(8):3308-3331. doi: 10.1093/molbev/msab105. Mol Biol Evol. 2021. PMID: 33871629 Free PMC article.
-
Genomic conservation of erythropoietic microRNAs (erythromiRs) in white-blooded Antarctic icefish.Mar Genomics. 2016 Dec;30:27-34. doi: 10.1016/j.margen.2016.04.013. Epub 2016 May 14. Mar Genomics. 2016. PMID: 27189439 Free PMC article.
-
The long noncoding RNA TUG1 regulates blood-tumor barrier permeability by targeting miR-144.Oncotarget. 2015 Aug 14;6(23):19759-79. doi: 10.18632/oncotarget.4331. Oncotarget. 2015. PMID: 26078353 Free PMC article.
References
-
- Broudy V.C., Lin N.L., Priestley G.V., Nocka K., Wolf N.S. 1996. Interaction of stem cell factor and its receptor c-kit mediates lodgment and acute expansion of hematopoietic cells in the murine spleen. Blood. 88:75–81 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

