Carboxylated N-glycans on RAGE promote S100A12 binding and signaling
- PMID: 20512925
- PMCID: PMC2879712
- DOI: 10.1002/jcb.22575
Carboxylated N-glycans on RAGE promote S100A12 binding and signaling
Erratum in
- J Cell Biochem. 2010 Sep 1;111(1):248
Abstract
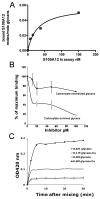
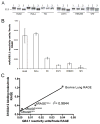
The receptor for advanced glycation end products (RAGE) is a signaling receptor protein of the immunoglobulin superfamily implicated in multiple pathologies. It binds a diverse repertoire of ligands, but the structural basis for the interaction of different ligands is not well understood. We earlier showed that carboxylated glycans on the V-domain of RAGE promote the binding of HMGB1 and S100A8/A9. Here we study the role of these glycans on the binding and intracellular signaling mediated by another RAGE ligand, S100A12. S100A12 binds carboxylated glycans, and a subpopulation of RAGE enriched for carboxylated glycans shows more than 10-fold higher binding potential for S100A12 than total RAGE. When expressed in mammalian cells, RAGE is modified by complex glycans predominantly at the first glycosylation site (N25IT) that retains S100A12 binding. Glycosylation of RAGE and maximum binding sites for S100A12 on RAGE are also cell type dependent. Carboxylated glycan-enriched population of RAGE forms higher order multimeric complexes with S100A12, and this ability to multimerize is reduced upon deglycosylation or by using non-glycosylated sRAGE expressed in E. coli. mAbGB3.1, an antibody against carboxylated glycans, blocks S100A12-mediated NF-kappaB signaling in HeLa cells expressing full-length RAGE. These results demonstrate that carboxylated N-glycans on RAGE enhance binding potential and promote receptor clustering and subsequent signaling events following oligomeric S100A12 binding.
(c) 2010 Wiley-Liss, Inc.
Figures
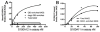


Similar articles
-
RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis.Carcinogenesis. 2008 Oct;29(10):2035-43. doi: 10.1093/carcin/bgn188. Epub 2008 Aug 9. Carcinogenesis. 2008. PMID: 18689872 Free PMC article.
-
N -Glycans on the receptor for advanced glycation end products influence amphoterin binding and neurite outgrowth.J Neurochem. 2002 Mar;80(6):998-1008. doi: 10.1046/j.0022-3042.2002.00796.x. J Neurochem. 2002. PMID: 11953450
-
Blocking the Interactions between Calcium-Bound S100A12 Protein and the V Domain of RAGE Using Tranilast.PLoS One. 2016 Sep 6;11(9):e0162000. doi: 10.1371/journal.pone.0162000. eCollection 2016. PLoS One. 2016. PMID: 27598566 Free PMC article.
-
RAGE during infectious diseases.Front Biosci (Schol Ed). 2011 Jun 1;3(3):1119-32. doi: 10.2741/215. Front Biosci (Schol Ed). 2011. PMID: 21622260 Review.
-
S100A12 and the S100/Calgranulins: Emerging Biomarkers for Atherosclerosis and Possibly Therapeutic Targets.Arterioscler Thromb Vasc Biol. 2015 Dec;35(12):2496-507. doi: 10.1161/ATVBAHA.115.302072. Epub 2015 Oct 29. Arterioscler Thromb Vasc Biol. 2015. PMID: 26515415 Free PMC article. Review.
Cited by
-
Tissue S100/calgranulin expression and blood neutrophil-to-lymphocyte ratio (NLR) in dogs with lower urinary tract urothelial carcinoma.BMC Vet Res. 2022 Nov 21;18(1):412. doi: 10.1186/s12917-022-03513-z. BMC Vet Res. 2022. PMID: 36411489 Free PMC article.
-
The N-glycoform of sRAGE is the key determinant for its therapeutic efficacy to attenuate injury-elicited arterial inflammation and neointimal growth.J Mol Med (Berl). 2013 Dec;91(12):1369-81. doi: 10.1007/s00109-013-1091-4. Epub 2013 Oct 17. J Mol Med (Berl). 2013. PMID: 24132651 Free PMC article.
-
Tranilast Blocks the Interaction between the Protein S100A11 and Receptor for Advanced Glycation End Products (RAGE) V Domain and Inhibits Cell Proliferation.J Biol Chem. 2016 Jul 1;291(27):14300-14310. doi: 10.1074/jbc.M116.722215. Epub 2016 May 12. J Biol Chem. 2016. PMID: 27226584 Free PMC article.
-
Chronic sustained inflammation links to left ventricular hypertrophy and aortic valve sclerosis: a new link between S100/RAGE and FGF23.Inflamm Cell Signal. 2014;1(5):e279. doi: 10.14800/ics.279. Inflamm Cell Signal. 2014. PMID: 26082935 Free PMC article.
-
Impact of Advanced Glycation End products (AGEs) and its receptor (RAGE) on cancer metabolic signaling pathways and its progression.Glycoconj J. 2021 Dec;38(6):717-734. doi: 10.1007/s10719-021-10031-x. Epub 2022 Jan 22. Glycoconj J. 2021. PMID: 35064413 Review.
References
-
- Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, Stern DM, Nawroth PP. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83:876–86. - PubMed
-
- Bierhaus A, Nawroth PP. Multiple levels of regulation determine the role of the receptor for AGE (RAGE) as common soil in inflammation, immune responses and diabetes mellitus and its complications. Diabetologia. 2009;52:2251–63. - PubMed
-
- Bierhaus A, Schiekofer S, Schwaninger M, Andrassy M, Humpert PM, Chen J, Hong M, Luther T, Henle T, Kloting I, Morcos M, Hofmann M, Tritschler H, Weigle B, Kasper M, Smith M, Perry G, Schmidt AM, Stern DM, Haring HU, Schleicher E, Nawroth PP. Diabetes-associated sustained activation of the transcription factor nuclear factor-kappaB. Diabetes. 2001;50:2792–808. - PubMed
-
- Brewer CF, Miceli MC, Baum LG. Clusters, bundles, arrays and lattices: novel mechanisms for lectin-saccharide-mediated cellular interactions. Curr Opin Struct Biol. 2002;12:616–23. - PubMed
-
- Cairo CW, Gestwicki JE, Kanai M, Kiessling LL. Control of multivalent interactions by binding epitope density. J Am Chem Soc. 2002;124:1615–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

