Relationship between nucleosome positioning and DNA methylation
- PMID: 20512117
- PMCID: PMC2964354
- DOI: 10.1038/nature09147
Relationship between nucleosome positioning and DNA methylation
Abstract
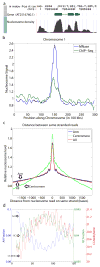
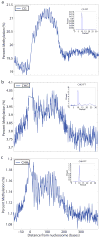
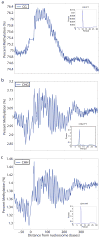
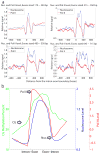
Nucleosomes compact and regulate access to DNA in the nucleus, and are composed of approximately 147 bases of DNA wrapped around a histone octamer. Here we report a genome-wide nucleosome positioning analysis of Arabidopsis thaliana using massively parallel sequencing of mononucleosomes. By combining this data with profiles of DNA methylation at single base resolution, we identified 10-base periodicities in the DNA methylation status of nucleosome-bound DNA and found that nucleosomal DNA was more highly methylated than flanking DNA. These results indicate that nucleosome positioning influences DNA methylation patterning throughout the genome and that DNA methyltransferases preferentially target nucleosome-bound DNA. We also observed similar trends in human nucleosomal DNA, indicating that the relationships between nucleosomes and DNA methyltransferases are conserved. Finally, as has been observed in animals, nucleosomes were highly enriched on exons, and preferentially positioned at intron-exon and exon-intron boundaries. RNA polymerase II (Pol II) was also enriched on exons relative to introns, consistent with the hypothesis that nucleosome positioning regulates Pol II processivity. DNA methylation is also enriched on exons, consistent with the targeting of DNA methylation to nucleosomes, and suggesting a role for DNA methylation in exon definition.
Figures
Similar articles
-
Nucleosome positioning, nucleosome spacing and the nucleosome code.J Biomol Struct Dyn. 2010 Jun;27(6):781-93. doi: 10.1080/073911010010524945. J Biomol Struct Dyn. 2010. PMID: 20232933 Free PMC article.
-
Genome-wide chromatin mapping with size resolution reveals a dynamic sub-nucleosomal landscape in Arabidopsis.PLoS Genet. 2017 Sep 13;13(9):e1006988. doi: 10.1371/journal.pgen.1006988. eCollection 2017 Sep. PLoS Genet. 2017. PMID: 28902852 Free PMC article.
-
Characterization of Dnmt1 Binding and DNA Methylation on Nucleosomes and Nucleosomal Arrays.PLoS One. 2015 Oct 23;10(10):e0140076. doi: 10.1371/journal.pone.0140076. eCollection 2015. PLoS One. 2015. PMID: 26496704 Free PMC article.
-
A structural perspective on the where, how, why, and what of nucleosome positioning.J Biomol Struct Dyn. 2010 Jun;27(6):803-20. doi: 10.1080/07391102.2010.10508585. J Biomol Struct Dyn. 2010. PMID: 20232935 Review.
-
Nucleosome positioning in yeasts: methods, maps, and mechanisms.Chromosoma. 2015 Jun;124(2):131-51. doi: 10.1007/s00412-014-0501-x. Epub 2014 Dec 23. Chromosoma. 2015. PMID: 25529773 Review.
Cited by
-
Gene promoters show chromosome-specificity and reveal chromosome territories in humans.BMC Genomics. 2013 Apr 24;14:278. doi: 10.1186/1471-2164-14-278. BMC Genomics. 2013. PMID: 23617842 Free PMC article.
-
Genome-wide DNA Methylation Signatures Are Determined by DNMT3A/B Sequence Preferences.Biochemistry. 2020 Jul 14;59(27):2541-2550. doi: 10.1021/acs.biochem.0c00339. Epub 2020 Jun 28. Biochemistry. 2020. PMID: 32543182 Free PMC article.
-
N6-methyldeoxyadenosine marks active transcription start sites in Chlamydomonas.Cell. 2015 May 7;161(4):879-892. doi: 10.1016/j.cell.2015.04.010. Epub 2015 Apr 30. Cell. 2015. PMID: 25936837 Free PMC article.
-
Chromatin modifications associated with diabetes.J Cardiovasc Transl Res. 2012 Aug;5(4):399-412. doi: 10.1007/s12265-012-9380-9. Epub 2012 May 26. J Cardiovasc Transl Res. 2012. PMID: 22639343 Review.
-
Transcription-coupled eviction of histones H2A/H2B governs V(D)J recombination.EMBO J. 2013 May 15;32(10):1381-92. doi: 10.1038/emboj.2013.42. Epub 2013 Mar 5. EMBO J. 2013. PMID: 23463099 Free PMC article.
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. - PubMed
-
- Kornberg RD, Lorch Y. Twenty–five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. - PubMed
-
- Albert I, et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–6. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

