Integrative structure modeling of macromolecular assemblies from proteomics data
- PMID: 20507923
- PMCID: PMC2938050
- DOI: 10.1074/mcp.R110.000067
Integrative structure modeling of macromolecular assemblies from proteomics data
Abstract
Proteomics techniques have been used to generate comprehensive lists of protein interactions in a number of species. However, relatively little is known about how these interactions result in functional multiprotein complexes. This gap can be bridged by combining data from proteomics experiments with data from established structure determination techniques. Correspondingly, integrative computational methods are being developed to provide descriptions of protein complexes at varying levels of accuracy and resolution, ranging from complex compositions to detailed atomic structures.
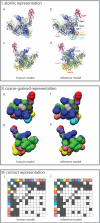
Figures


Similar articles
-
Structure determination of transient transcription complexes.Biochem Soc Trans. 2016 Aug 15;44(4):1177-82. doi: 10.1042/BST20160079. Biochem Soc Trans. 2016. PMID: 27528766 Review.
-
Modeling of proteins and their assemblies with the integrative modeling platform.Methods Mol Biol. 2011;781:377-97. doi: 10.1007/978-1-61779-276-2_19. Methods Mol Biol. 2011. PMID: 21877292
-
Subunit architecture of intact protein complexes from mass spectrometry and homology modeling.Acc Chem Res. 2008 May;41(5):617-27. doi: 10.1021/ar700218q. Epub 2008 Mar 4. Acc Chem Res. 2008. PMID: 18314965
-
Conserved RNA polymerase II initiation complex structure.Curr Opin Struct Biol. 2017 Dec;47:17-22. doi: 10.1016/j.sbi.2017.03.013. Epub 2017 Apr 22. Curr Opin Struct Biol. 2017. PMID: 28437704 Review.
-
Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies.PLoS Biol. 2012 Jan;10(1):e1001244. doi: 10.1371/journal.pbio.1001244. Epub 2012 Jan 17. PLoS Biol. 2012. PMID: 22272186 Free PMC article.
Cited by
-
UCSF Chimera, MODELLER, and IMP: an integrated modeling system.J Struct Biol. 2012 Sep;179(3):269-78. doi: 10.1016/j.jsb.2011.09.006. Epub 2011 Sep 22. J Struct Biol. 2012. PMID: 21963794 Free PMC article.
-
Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway.Mol Cell Proteomics. 2014 Nov;13(11):2855-70. doi: 10.1074/mcp.M114.039388. Epub 2014 Jul 29. Mol Cell Proteomics. 2014. PMID: 25073740 Free PMC article.
-
Systems Proteomics View of the Endogenous Human Claudin Protein Family.J Proteome Res. 2016 Feb 5;15(2):339-59. doi: 10.1021/acs.jproteome.5b00769. Epub 2016 Jan 12. J Proteome Res. 2016. PMID: 26680015 Free PMC article. Review.
-
A critical assessment of information-guided protein-protein docking predictions.Mol Cell Proteomics. 2013 Mar;12(3):679-86. doi: 10.1074/mcp.M112.020198. Epub 2012 Dec 13. Mol Cell Proteomics. 2013. PMID: 23242549 Free PMC article.
-
Toward an integrated structural model of the 26S proteasome.Mol Cell Proteomics. 2010 Aug;9(8):1666-77. doi: 10.1074/mcp.R000002-MCP201. Epub 2010 May 13. Mol Cell Proteomics. 2010. PMID: 20467039 Free PMC article.
References
-
- Alberts B. (1998) The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell 92, 291–294 - PubMed
-
- Abbott A. (2002) Proteomics: the society of proteins. Nature 417, 894–896 - PubMed
-
- Schmeing T. M., Ramakrishnan V. (2009) What recent ribosome structures have revealed about the mechanism of translation. Nature 461, 1234–1242 - PubMed
-
- Allen G. S., Frank J. (2007) Structural insights on the translation initiation complex: ghosts of a universal initiation complex. Mol. Microbiol. 63, 941–950 - PubMed
-
- Horwich A. L., Fenton W. A. (2009) Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 42, 83–116 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

