RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases
- PMID: 20497334
- PMCID: PMC2936116
- DOI: 10.1111/j.1365-2958.2010.07208.x
RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases
Abstract
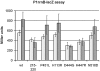
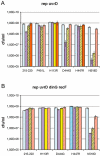
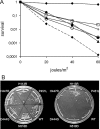
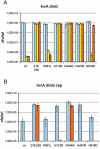
We observed that cells lacking Rep and UvrD, two replication accessory helicases, and the recombination protein RecF are cryo-sensitive on rich medium. We isolated five mutations that suppress this Luria-Bertani (LB)-cryo-sensitivity and show that they map in the genes encoding the RNA polymerase subunits RpoB and RpoC. These rpoB (D444G, H447R and N518D) and rpoC mutants (H113R and P451L) were characterized. rpoB(H447R) and rpoB(D444G) prevent activation of the Prrn core promoter in rich medium, but only rpoB(H447R) also suppresses the auxotrophy of a relA spoT mutant (stringent-like phenotype). rpoC(H113R) suppresses the thermo-sensitivity of a greA greB mutant, suggesting that it destabilizes stalled elongation complexes. All mutations but rpoC(P451L) prevent R-loop formation. We propose that these rpo mutations allow replication in the absence of Rep and UvrD by destabilizing RNA Pol upon replication-transcription collisions. In a RecF(+) context, they improve growth of rep uvrD cells only if DinG is present, supporting the hypothesis that Rep, UvrD and DinG facilitate progression of the replication fork across transcribed sequences. They rescue rep uvrD dinG recF cells, indicating that in a recF mutant replication forks arrested by unstable transcription complexes can restart without any of the three known replication accessory helicases Rep, UvrD and DinG.
Figures
Similar articles
-
The helicases DinG, Rep and UvrD cooperate to promote replication across transcription units in vivo.EMBO J. 2010 Jan 6;29(1):145-57. doi: 10.1038/emboj.2009.308. Epub 2009 Oct 22. EMBO J. 2010. PMID: 19851282 Free PMC article.
-
Direct removal of RNA polymerase barriers to replication by accessory replicative helicases.Nucleic Acids Res. 2019 Jun 4;47(10):5100-5113. doi: 10.1093/nar/gkz170. Nucleic Acids Res. 2019. PMID: 30869136 Free PMC article.
-
UvrD and UvrD252 counteract RecQ, RecJ, and RecFOR in a rep mutant of Escherichia coli.J Bacteriol. 2008 Sep;190(17):5995-6001. doi: 10.1128/JB.00620-08. Epub 2008 Jun 20. J Bacteriol. 2008. PMID: 18567657 Free PMC article.
-
Accessory replicative helicases and the replication of protein-bound DNA.J Mol Biol. 2014 Dec 12;426(24):3917-3928. doi: 10.1016/j.jmb.2014.10.001. Epub 2014 Oct 13. J Mol Biol. 2014. PMID: 25308339 Review.
-
What happens when replication and transcription complexes collide?Cell Cycle. 2010 Jul 1;9(13):2537-43. doi: 10.4161/cc.9.13.12122. Cell Cycle. 2010. PMID: 20581460 Free PMC article. Review.
Cited by
-
Modulation of DNA damage tolerance in Escherichia coli recG and ruv strains by mutations affecting PriB, the ribosome and RNA polymerase.Mol Microbiol. 2012 Nov;86(3):675-91. doi: 10.1111/mmi.12010. Epub 2012 Sep 7. Mol Microbiol. 2012. PMID: 22957744 Free PMC article.
-
RadD Contributes to R-Loop Avoidance in Sub-MIC Tobramycin.mBio. 2019 Jul 2;10(4):e01173-19. doi: 10.1128/mBio.01173-19. mBio. 2019. PMID: 31266870 Free PMC article.
-
The Balance between Recombination Enzymes and Accessory Replicative Helicases in Facilitating Genome Duplication.Genes (Basel). 2016 Jul 29;7(8):42. doi: 10.3390/genes7080042. Genes (Basel). 2016. PMID: 27483323 Free PMC article.
-
Acid-adapted strains of Escherichia coli K-12 obtained by experimental evolution.Appl Environ Microbiol. 2015 Mar;81(6):1932-41. doi: 10.1128/AEM.03494-14. Epub 2015 Jan 2. Appl Environ Microbiol. 2015. PMID: 25556191 Free PMC article.
-
Cellular characterization of the primosome and rep helicase in processing and restoration of replication following arrest by UV-induced DNA damage in Escherichia coli.J Bacteriol. 2012 Aug;194(15):3977-86. doi: 10.1128/JB.00290-12. Epub 2012 May 25. J Bacteriol. 2012. PMID: 22636770 Free PMC article.
References
-
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
-
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001a;305:689–702. - PubMed
-
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001b;305:673–688. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

