ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids
- PMID: 20478830
- PMCID: PMC2896094
- DOI: 10.1093/nar/gkq399
ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids
Abstract
It is informative to detect highly conserved positions in proteins and nucleic acid sequence/structure since they are often indicative of structural and/or functional importance. ConSurf (http://consurf.tau.ac.il) and ConSeq (http://conseq.tau.ac.il) are two well-established web servers for calculating the evolutionary conservation of amino acid positions in proteins using an empirical Bayesian inference, starting from protein structure and sequence, respectively. Here, we present the new version of the ConSurf web server that combines the two independent servers, providing an easier and more intuitive step-by-step interface, while offering the user more flexibility during the process. In addition, the new version of ConSurf calculates the evolutionary rates for nucleic acid sequences. The new version is freely available at: http://consurf.tau.ac.il/.
Figures
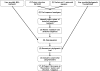

Similar articles
-
ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules.Nucleic Acids Res. 2016 Jul 8;44(W1):W344-50. doi: 10.1093/nar/gkw408. Epub 2016 May 10. Nucleic Acids Res. 2016. PMID: 27166375 Free PMC article.
-
ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures.Nucleic Acids Res. 2005 Jul 1;33(Web Server issue):W299-302. doi: 10.1093/nar/gki370. Nucleic Acids Res. 2005. PMID: 15980475 Free PMC article.
-
The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures.Nucleic Acids Res. 2009 Jan;37(Database issue):D323-7. doi: 10.1093/nar/gkn822. Epub 2008 Oct 29. Nucleic Acids Res. 2009. PMID: 18971256 Free PMC article.
-
The ConSurf-HSSP database: the mapping of evolutionary conservation among homologs onto PDB structures.Proteins. 2005 Feb 15;58(3):610-7. doi: 10.1002/prot.20305. Proteins. 2005. PMID: 15614759
-
Wenxiang 3.0: Evolutionary Visualization of α, π, and 3/10 Helices.Evol Bioinform Online. 2022 May 31;18:11769343221101014. doi: 10.1177/11769343221101014. eCollection 2022. Evol Bioinform Online. 2022. PMID: 35668741 Free PMC article. Review.
Cited by
-
Sequence search and analysis of gene products containing RNA recognition motifs in the human genome.BMC Genomics. 2014 Dec 22;15(1):1159. doi: 10.1186/1471-2164-15-1159. BMC Genomics. 2014. PMID: 25534245 Free PMC article.
-
The organization of a CSN5-containing subcomplex of the COP9 signalosome.J Biol Chem. 2012 Dec 7;287(50):42031-41. doi: 10.1074/jbc.M112.387977. Epub 2012 Oct 18. J Biol Chem. 2012. PMID: 23086934 Free PMC article.
-
eIF4E-binding protein regulation of mRNAs with differential 5'-UTR secondary structure: a polyelectrostatic model for a component of protein-mRNA interactions.Nucleic Acids Res. 2012 Sep;40(16):7666-75. doi: 10.1093/nar/gks511. Epub 2012 Jun 20. Nucleic Acids Res. 2012. PMID: 22718971 Free PMC article.
-
CLIPS-1D: analysis of multiple sequence alignments to deduce for residue-positions a role in catalysis, ligand-binding, or protein structure.BMC Bioinformatics. 2012 Apr 5;13:55. doi: 10.1186/1471-2105-13-55. BMC Bioinformatics. 2012. PMID: 22480135 Free PMC article.
-
Structure of a Bud6/Actin Complex Reveals a Novel WH2-like Actin Monomer Recruitment Motif.Structure. 2015 Aug 4;23(8):1492-1499. doi: 10.1016/j.str.2015.05.015. Epub 2015 Jun 25. Structure. 2015. PMID: 26118535 Free PMC article.
References
-
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. - PubMed
-
- Berezin C, Glaser F, Rosenberg J, Paz I, Pupko T, Fariselli P, Casadio R, Ben-Tal N. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics. 2004;20:1322–1324. - PubMed
-
- Mayrose I, Graur D, Ben-Tal N, Pupko T. Comparison of site-specific rate-inference methods for protein sequences: empirical Bayesian methods are superior. Mol. Biol. Evol. 2004;21:1781–1791. - PubMed
-
- Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl 1):S71–S77. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

