B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast
- PMID: 20463882
- PMCID: PMC2865516
- DOI: 10.1371/journal.pgen.1000935
B-cyclin/CDKs regulate mitotic spindle assembly by phosphorylating kinesins-5 in budding yeast
Abstract
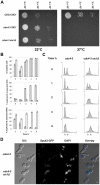
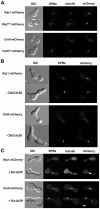
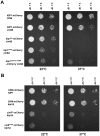
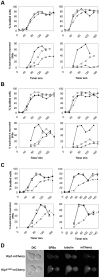
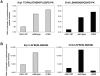
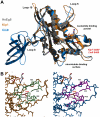
Although it has been known for many years that B-cyclin/CDK complexes regulate the assembly of the mitotic spindle and entry into mitosis, the full complement of relevant CDK targets has not been identified. It has previously been shown in a variety of model systems that B-type cyclin/CDK complexes, kinesin-5 motors, and the SCF(Cdc4) ubiquitin ligase are required for the separation of spindle poles and assembly of a bipolar spindle. It has been suggested that, in budding yeast, B-type cyclin/CDK (Clb/Cdc28) complexes promote spindle pole separation by inhibiting the degradation of the kinesins-5 Kip1 and Cin8 by the anaphase-promoting complex (APC(Cdh1)). We have determined, however, that the Kip1 and Cin8 proteins are present at wild-type levels in the absence of Clb/Cdc28 kinase activity. Here, we show that Kip1 and Cin8 are in vitro targets of Clb2/Cdc28 and that the mutation of conserved CDK phosphorylation sites on Kip1 inhibits spindle pole separation without affecting the protein's in vivo localization or abundance. Mass spectrometry analysis confirms that two CDK sites in the tail domain of Kip1 are phosphorylated in vivo. In addition, we have determined that Sic1, a Clb/Cdc28-specific inhibitor, is the SCF(Cdc4) target that inhibits spindle pole separation in cells lacking functional Cdc4. Based on these findings, we propose that Clb/Cdc28 drives spindle pole separation by direct phosphorylation of kinesin-5 motors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
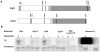

Similar articles
-
Cdk1 regulates centrosome separation by restraining proteolysis of microtubule-associated proteins.EMBO J. 2006 Jun 7;25(11):2551-63. doi: 10.1038/sj.emboj.7601136. Epub 2006 May 11. EMBO J. 2006. PMID: 16688214 Free PMC article.
-
Spindle pole body separation in Saccharomyces cerevisiae requires dephosphorylation of the tyrosine 19 residue of Cdc28.Mol Cell Biol. 1996 Nov;16(11):6385-97. doi: 10.1128/MCB.16.11.6385. Mol Cell Biol. 1996. PMID: 8887667 Free PMC article.
-
Inactivation of Cdh1 by synergistic action of Cdk1 and polo kinase is necessary for proper assembly of the mitotic spindle.Nat Cell Biol. 2008 Jun;10(6):665-75. doi: 10.1038/ncb1729. Epub 2008 May 25. Nat Cell Biol. 2008. PMID: 18500339 Free PMC article.
-
Mitotic motors in Saccharomyces cerevisiae.Biochim Biophys Acta. 2000 Mar 17;1496(1):99-116. doi: 10.1016/s0167-4889(00)00012-4. Biochim Biophys Acta. 2000. PMID: 10722880 Review.
-
Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae.Microbiol Mol Biol Rev. 1998 Dec;62(4):1191-243. doi: 10.1128/MMBR.62.4.1191-1243.1998. Microbiol Mol Biol Rev. 1998. PMID: 9841670 Free PMC article. Review.
Cited by
-
Mechanisms by Which Kinesin-5 Motors Perform Their Multiple Intracellular Functions.Int J Mol Sci. 2021 Jun 15;22(12):6420. doi: 10.3390/ijms22126420. Int J Mol Sci. 2021. PMID: 34203964 Free PMC article. Review.
-
Synthetic-Evolution Reveals Narrow Paths to Regulation of the Saccharomyces cerevisiae Mitotic Kinesin-5 Cin8.Int J Biol Sci. 2019 May 2;15(6):1125-1138. doi: 10.7150/ijbs.30543. eCollection 2019. Int J Biol Sci. 2019. PMID: 31223274 Free PMC article.
-
Mechanism and regulation of kinesin-5, an essential motor for the mitotic spindle.Biol Cell. 2014 Jan;106(1):1-12. doi: 10.1111/boc.201300054. Epub 2013 Nov 26. Biol Cell. 2014. PMID: 24125467 Free PMC article. Review.
-
Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain.Sci Rep. 2016 May 24;6:25597. doi: 10.1038/srep25597. Sci Rep. 2016. PMID: 27216310 Free PMC article.
-
Centrosome Remodelling in Evolution.Cells. 2018 Jul 6;7(7):71. doi: 10.3390/cells7070071. Cells. 2018. PMID: 29986477 Free PMC article. Review.
References
-
- Morgan DO. Cyclin-dependent kinases: Engines, clocks, and microprocessors. Annual Review of Cell and Developmental Biology. 1997;13:261–291. - PubMed
-
- Murray AW. Recycling the cell cycle: Cyclins revisited. Cell. 2004;116:221–234. - PubMed
-
- Haase SB, Winey M, Reed SI. Multi-step control of spindle pole body duplication by cyclin-dependent kinase. Nature Cell Biology. 2001;3:38–42. - PubMed
-
- Adams IR, Kilmartin JV. Spindle pole body duplication: a model for centrosome duplication? Trends in Cell Biology. 2000;10:329–335. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

