A survey of sequence alignment algorithms for next-generation sequencing
- PMID: 20460430
- PMCID: PMC2943993
- DOI: 10.1093/bib/bbq015
A survey of sequence alignment algorithms for next-generation sequencing
Abstract
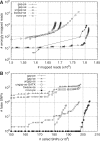
Rapidly evolving sequencing technologies produce data on an unparalleled scale. A central challenge to the analysis of this data is sequence alignment, whereby sequence reads must be compared to a reference. A wide variety of alignment algorithms and software have been subsequently developed over the past two years. In this article, we will systematically review the current development of these algorithms and introduce their practical applications on different types of experimental data. We come to the conclusion that short-read alignment is no longer the bottleneck of data analyses. We also consider future development of alignment algorithms with respect to emerging long sequence reads and the prospect of cloud computing.
Figures
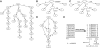


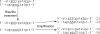
Similar articles
-
Long Read Alignment with Parallel MapReduce Cloud Platform.Biomed Res Int. 2015;2015:807407. doi: 10.1155/2015/807407. Epub 2015 Dec 29. Biomed Res Int. 2015. PMID: 26839887 Free PMC article.
-
Alignment of Next-Generation Sequencing Reads.Annu Rev Genomics Hum Genet. 2015;16:133-51. doi: 10.1146/annurev-genom-090413-025358. Epub 2015 May 4. Annu Rev Genomics Hum Genet. 2015. PMID: 25939052 Review.
-
Improved variant discovery through local re-alignment of short-read next-generation sequencing data using SRMA.Genome Biol. 2010;11(10):R99. doi: 10.1186/gb-2010-11-10-r99. Epub 2010 Oct 8. Genome Biol. 2010. PMID: 20932289 Free PMC article.
-
RandAL: a randomized approach to aligning DNA sequences to reference genomes.BMC Genomics. 2014;15 Suppl 5(Suppl 5):S2. doi: 10.1186/1471-2164-15-S5-S2. Epub 2014 Jul 14. BMC Genomics. 2014. PMID: 25081493 Free PMC article.
-
Sense from sequence reads: methods for alignment and assembly.Nat Methods. 2009 Nov;6(11 Suppl):S6-S12. doi: 10.1038/nmeth.1376. Nat Methods. 2009. PMID: 19844229 Review.
Cited by
-
The Ensembl gene annotation system.Database (Oxford). 2016 Jun 23;2016:baw093. doi: 10.1093/database/baw093. Print 2016. Database (Oxford). 2016. PMID: 27337980 Free PMC article.
-
Indel-tolerant read mapping with trinucleotide frequencies using cache-oblivious kd-trees.Bioinformatics. 2012 Sep 15;28(18):i325-i332. doi: 10.1093/bioinformatics/bts380. Bioinformatics. 2012. PMID: 22962448 Free PMC article.
-
Cardiovascular genomics: a biomarker identification pipeline.IEEE Trans Inf Technol Biomed. 2012 Sep;16(5):809-22. doi: 10.1109/TITB.2012.2199570. Epub 2012 May 16. IEEE Trans Inf Technol Biomed. 2012. PMID: 22614726 Free PMC article. Review.
-
Computational Methods for Strain-Level Microbial Detection in Colony and Metagenome Sequencing Data.Front Microbiol. 2020 Aug 18;11:1925. doi: 10.3389/fmicb.2020.01925. eCollection 2020. Front Microbiol. 2020. PMID: 33013732 Free PMC article. Review.
-
CloudMap: a cloud-based pipeline for analysis of mutant genome sequences.Genetics. 2012 Dec;192(4):1249-69. doi: 10.1534/genetics.112.144204. Epub 2012 Oct 10. Genetics. 2012. PMID: 23051646 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

