KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation
- PMID: 20457893
- PMCID: PMC2906833
- DOI: 10.1073/pnas.1000401107
KDM8, a H3K36me2 histone demethylase that acts in the cyclin A1 coding region to regulate cancer cell proliferation
Abstract
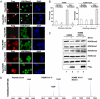
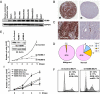
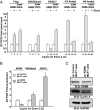
Localized chromatin modifications of histone tails play an important role in regulating gene transcription, and aberration of these processes leads to carcinogenesis. Methylated histone lysine residues, a key player in chromatin remodeling, are demethylated by the JmjC class of enzymes. Here we show that JMJD5 (now renamed KDM8), a JmjC family member, demethylates H3K36me2 and is required for cell cycle progression. Chromatin immunoprecipitation assays applied to human genome tiling arrays in conjunction with RNA microarray revealed that KDM8 occupies the coding region of cyclin A1 and directly regulates transcription. Mechanistic analyses showed that KDM8 functioned as a transcriptional activator by inhibiting HDAC recruitment via demethylation of H3K36me2, an epigenetic repressive mark. Tumor array experiments revealed KDM8 is overexpressed in several types of cancer. In addition, loss-of-function studies in MCF7 cells leads to cell cycle arrest. These studies identified KDM8 as an important cell cycle regulator.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Epigenetic silencing of JMJD5 promotes the proliferation of hepatocellular carcinoma cells by down-regulating the transcription of CDKN1A 686.Oncotarget. 2016 Feb 9;7(6):6847-63. doi: 10.18632/oncotarget.6867. Oncotarget. 2016. PMID: 26760772 Free PMC article.
-
Jmjd5, an H3K36me2 histone demethylase, modulates embryonic cell proliferation through the regulation of Cdkn1a expression.Development. 2012 Feb;139(4):749-59. doi: 10.1242/dev.074138. Epub 2012 Jan 12. Development. 2012. PMID: 22241836
-
Identification and functional implication of nuclear localization signals in the N-terminal domain of JMJD5.Biochimie. 2013 Nov;95(11):2114-22. doi: 10.1016/j.biochi.2013.08.002. Epub 2013 Aug 12. Biochimie. 2013. PMID: 23948433
-
Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells.Cancer Sci. 2016 Sep;107(9):1187-92. doi: 10.1111/cas.13004. Epub 2016 Sep 1. Cancer Sci. 2016. PMID: 27375009 Free PMC article. Review.
-
Histone acetylation and the cell-cycle in cancer.Front Biosci. 2001 Apr 1;6:D610-29. doi: 10.2741/1wang1. Front Biosci. 2001. PMID: 11282573 Review.
Cited by
-
The emerging roles of histone demethylases in cancers.Cancer Metastasis Rev. 2024 Jun;43(2):795-821. doi: 10.1007/s10555-023-10160-9. Epub 2024 Jan 16. Cancer Metastasis Rev. 2024. PMID: 38227150 Review.
-
Breathing-in epigenetic change with vitamin C.EMBO Rep. 2013 Apr;14(4):337-46. doi: 10.1038/embor.2013.29. Epub 2013 Mar 15. EMBO Rep. 2013. PMID: 23492828 Free PMC article. Review.
-
Tumor hypoxia: From basic knowledge to therapeutic implications.Semin Cancer Biol. 2023 Jan;88:172-186. doi: 10.1016/j.semcancer.2022.12.011. Epub 2023 Jan 2. Semin Cancer Biol. 2023. PMID: 36603793 Free PMC article. Review.
-
JMJD5 inhibits lung cancer progression by facilitating EGFR proteasomal degradation.Cell Death Dis. 2023 Oct 9;14(10):657. doi: 10.1038/s41419-023-06194-0. Cell Death Dis. 2023. PMID: 37813845 Free PMC article.
-
To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families.Int J Mol Sci. 2024 Jun 24;25(13):6900. doi: 10.3390/ijms25136900. Int J Mol Sci. 2024. PMID: 39000010 Free PMC article. Review.
References
-
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. - PubMed
-
- Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. - PubMed
-
- Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. - PubMed
-
- Iwase S, et al. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. - PubMed
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

