Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells
- PMID: 20439434
- PMCID: PMC2861194
- DOI: 10.1101/gad.1886810
Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells
Abstract
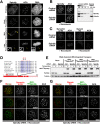
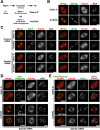
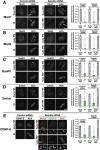
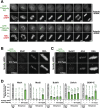
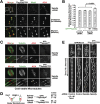
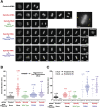
The spindle checkpoint generates a "wait anaphase" signal at unattached kinetochores to prevent premature anaphase onset. Kinetochore-localized dynein is thought to silence the checkpoint by transporting checkpoint proteins from microtubule-attached kinetochores to spindle poles. Throughout metazoans, dynein recruitment to kinetochores requires the protein Spindly. Here, we identify a conserved motif in Spindly that is essential for kinetochore targeting of dynein. Spindly motif mutants, expressed following depletion of endogenous Spindly, target normally to kinetochores but prevent dynein recruitment. Spindly depletion and Spindly motif mutants, despite their similar effects on kinetochore dynein, have opposite consequences on chromosome alignment and checkpoint silencing. Spindly depletion delays chromosome alignment, but Spindly motif mutants ameliorate this defect, indicating that Spindly has a dynein recruitment-independent role in alignment. In Spindly depletions, the checkpoint is silenced following delayed alignment by a kinetochore dynein-independent mechanism. In contrast, Spindly motif mutants are retained on microtubule-attached kinetochores along with checkpoint proteins, resulting in persistent checkpoint signaling. Thus, dynein-mediated removal of Spindly from microtubule-attached kinetochores, rather than poleward transport per se, is the critical reaction in checkpoint silencing. In the absence of Spindly, a second mechanism silences the checkpoint; this mechanism is likely evolutionarily ancient, as fungi and higher plants lack kinetochore dynein.
Figures
Similar articles
-
Molecular mechanism of dynein recruitment to kinetochores by the Rod-Zw10-Zwilch complex and Spindly.J Cell Biol. 2017 Apr 3;216(4):943-960. doi: 10.1083/jcb.201610108. Epub 2017 Mar 20. J Cell Biol. 2017. PMID: 28320824 Free PMC article.
-
Spindly switch controls anaphase: spindly and RZZ functions in chromosome attachment and mitotic checkpoint control.Cell Cycle. 2011 Feb 1;10(3):449-56. doi: 10.4161/cc.10.3.14759. Epub 2011 Feb 1. Cell Cycle. 2011. PMID: 21252629 Review.
-
Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore.J Cell Biol. 2007 Jun 18;177(6):1005-15. doi: 10.1083/jcb.200702062. J Cell Biol. 2007. PMID: 17576797 Free PMC article.
-
Dynein/Dynactin-mediated transport of kinetochore components off kinetochores and onto spindle poles induced by nordihydroguaiaretic acid.PLoS One. 2011 Jan 28;6(1):e16494. doi: 10.1371/journal.pone.0016494. PLoS One. 2011. PMID: 21305043 Free PMC article.
-
RZZ-SPINDLY-DYNEIN: you got to keep 'em separated.Cell Cycle. 2020 Jul;19(14):1716-1726. doi: 10.1080/15384101.2020.1780382. Epub 2020 Jun 16. Cell Cycle. 2020. PMID: 32544383 Free PMC article. Review.
Cited by
-
Cohesion fatigue explains why pharmacological inhibition of the APC/C induces a spindle checkpoint-dependent mitotic arrest.PLoS One. 2012;7(11):e49041. doi: 10.1371/journal.pone.0049041. Epub 2012 Nov 7. PLoS One. 2012. PMID: 23145059 Free PMC article.
-
Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores.J Cell Biol. 2013 Sep 2;202(5):735-46. doi: 10.1083/jcb.201304197. Epub 2013 Aug 26. J Cell Biol. 2013. PMID: 23979716 Free PMC article.
-
The Disordered Spindly C-terminus Interacts with RZZ Subunits ROD-1 and ZWL-1 in the Kinetochore through the Same Sites in C. Elegans.J Mol Biol. 2021 Feb 19;433(4):166812. doi: 10.1016/j.jmb.2021.166812. Epub 2021 Jan 13. J Mol Biol. 2021. PMID: 33450249 Free PMC article.
-
Involvement of the NF-κB signaling pathway in proliferation and invasion inhibited by Zwint-1 deficiency in Pancreatic Cancer Cells.J Cancer. 2020 Jul 25;11(19):5601-5611. doi: 10.7150/jca.46173. eCollection 2020. J Cancer. 2020. PMID: 32913455 Free PMC article.
-
Conformational transitions of the Spindly adaptor underlie its interaction with Dynein and Dynactin.J Cell Biol. 2022 Nov 7;221(11):e202206131. doi: 10.1083/jcb.202206131. Epub 2022 Sep 15. J Cell Biol. 2022. PMID: 36107127 Free PMC article.
References
-
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC 2001. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106: 83–93 - PubMed
-
- Buffin E, Lefebvre C, Huang J, Gagou ME, Karess R 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15: 856–861 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
