Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity
- PMID: 20413611
- PMCID: PMC2867212
- DOI: 10.1101/gad.1890410
Precise temporal control of the eye regulatory gene Pax6 via enhancer-binding site affinity
Abstract
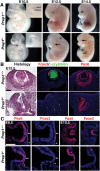
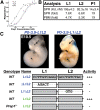
How transcription factors interpret the cis-regulatory logic encoded within enhancers to mediate quantitative changes in spatiotemporally restricted expression patterns during animal development is not well understood. Pax6 is a dosage-sensitive gene essential for eye development. Here, we identify the Prep1 (pKnox1) transcription factor as a critical dose-dependent upstream regulator of Pax6 expression during lens formation. We show that Prep1 activates the Pax6 lens enhancer by binding to two phylogenetically conserved lower-affinity DNA-binding sites. Finally, we describe a mechanism whereby Pax6 levels are determined by transcriptional synergy of Prep1 bound to the two sites, while timing of enhancer activation is determined by binding site affinity.
Figures


Similar articles
-
The functional role of the Meis/Prep-binding elements in Pax6 locus during pancreas and eye development.Dev Biol. 2012 Mar 1;363(1):320-9. doi: 10.1016/j.ydbio.2011.12.038. Epub 2012 Jan 3. Dev Biol. 2012. PMID: 22240097 Free PMC article.
-
PAX6 and SOX2-dependent regulation of the Sox2 enhancer N-3 involved in embryonic visual system development.Genes Cells. 2007 Sep;12(9):1049-61. doi: 10.1111/j.1365-2443.2007.01114.x. Genes Cells. 2007. PMID: 17825048
-
Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development.Nucleic Acids Res. 2015 Aug 18;43(14):6827-46. doi: 10.1093/nar/gkv589. Epub 2015 Jul 2. Nucleic Acids Res. 2015. PMID: 26138486 Free PMC article.
-
Interplay of Pax6 and SOX2 in lens development as a paradigm of genetic switch mechanisms for cell differentiation.Int J Dev Biol. 2004;48(8-9):819-27. doi: 10.1387/ijdb.041868hk. Int J Dev Biol. 2004. PMID: 15558474 Review.
-
Regulation of gene expression by Pax6 in ocular cells: a case of tissue-preferred expression of crystallins in lens.Int J Dev Biol. 2004;48(8-9):829-44. doi: 10.1387/ijdb.041866ac. Int J Dev Biol. 2004. PMID: 15558475 Free PMC article. Review.
Cited by
-
Why detailed model gene studies in higher eukaryotes are still necessary.Immunology. 2013 Jun;139(2):158-60. doi: 10.1111/imm.12066. Immunology. 2013. PMID: 23311893 Free PMC article. Review.
-
A model of spatially restricted transcription in opposing gradients of activators and repressors.Mol Syst Biol. 2012;8:614. doi: 10.1038/msb.2012.48. Mol Syst Biol. 2012. PMID: 23010997 Free PMC article.
-
Temporal Control of the TGF-β Signaling Network by Mouse ESC MicroRNA Targets of Different Affinities.Cell Rep. 2019 Nov 26;29(9):2702-2717.e7. doi: 10.1016/j.celrep.2019.10.109. Cell Rep. 2019. PMID: 31775039 Free PMC article.
-
DNA binding analysis of rare variants in homeodomains reveals homeodomain specificity-determining residues.Nat Commun. 2024 Apr 10;15(1):3110. doi: 10.1038/s41467-024-47396-0. Nat Commun. 2024. PMID: 38600112 Free PMC article.
-
The functional role of the Meis/Prep-binding elements in Pax6 locus during pancreas and eye development.Dev Biol. 2012 Mar 1;363(1):320-9. doi: 10.1016/j.ydbio.2011.12.038. Epub 2012 Jan 3. Dev Biol. 2012. PMID: 22240097 Free PMC article.
References
-
- Aota S, Nakajima N, Sakamoto R, Watanabe S, Ibaraki N, Okazaki K 2003. Pax6 autoregulation mediated by direct interaction of Pax6 protein with the head surface ectoderm-specific enhancer of the mouse Pax6 gene. Dev Biol 257: 1–13 - PubMed
-
- Arnosti DN, Kulkarni MM 2005. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
