Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism
- PMID: 20404320
- PMCID: PMC2898347
- DOI: 10.1074/jbc.M110.125674
Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism
Abstract
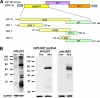
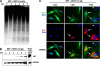
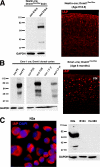
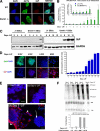
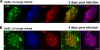
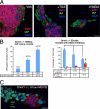
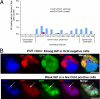
In defense of deleterious retrotransposition of intracisternal A particle (IAP) elements, IAP loci are heavily methylated and silenced in mouse somatic cells. To determine whether IAP is also repressed in pluripotent stem cells by DNA methylation, we examined IAP expression in demethylated mouse embryonic stem cells (mESCs) and epiblast-derived stem cells. Surprisingly, in demethylated ESC cultures carrying mutations of DNA methyltransferase I (Dnmt1), no IAP transcripts and proteins are detectable in undifferentiated Oct4(+) ESCs. In contrast, approximately 3.6% of IAP-positive cells are detected in Oct4(-) Dnmt1(-/-) cells, suggesting that the previously observed increase in IAP transcripts in the population of Dnmt1(-/-) ESCs could be accounted for by this subset of Oct4(-) Dnmt1(-/-) ESCs undergoing spontaneous differentiation. Consistent with this possibility, a dramatic increase of IAP mRNA (>100-fold) and protein expression was observed in Dnmt1(-/-) ESC cultures upon induction of differentiation through the withdrawal of leukemia-inhibitory factor for 6 or more days. Interestingly, both mRNAs and proteins of IAP can be readily detected in demethylated Oct4(+) epiblast-derived stem cells as well as differentiated mouse embryo fibroblasts, neurons, and glia upon conditional Dnmt1 gene deletion. These data suggest that mESCs are a unique stem cell type possessing a DNA methylation-independent IAP repression mechanism. This methylation-independent mechanism does not involve Dicer-mediated action of microRNAs or RNA interference because IAP expression remains repressed in Dnmt1(-/-); Dicer(-/-) double mutant ESCs. We suggest that mESCs possess a unique DNA methylation-independent mechanism to silence retrotransposons to safeguard genome stability while undergoing rapid cell proliferation for self-renewal.
Figures
Similar articles
-
DNMT1 can induce primary germ layer differentiation through de novo DNA methylation.Genes Cells. 2024 Jul;29(7):549-566. doi: 10.1111/gtc.13130. Epub 2024 May 29. Genes Cells. 2024. PMID: 38811355 Free PMC article.
-
Dnmt1 expression in pre- and postimplantation embryogenesis and the maintenance of IAP silencing.Mol Cell Biol. 2004 Feb;24(4):1640-8. doi: 10.1128/MCB.24.4.1640-1648.2004. Mol Cell Biol. 2004. PMID: 14749379 Free PMC article.
-
DNMT1 regulates the timing of DNA methylation by DNMT3 in an enzymatic activity-dependent manner in mouse embryonic stem cells.PLoS One. 2022 Jan 5;17(1):e0262277. doi: 10.1371/journal.pone.0262277. eCollection 2022. PLoS One. 2022. PMID: 34986190 Free PMC article.
-
DNA methylation in embryonic stem cells.J Cell Biochem. 2010 Jan 1;109(1):1-6. doi: 10.1002/jcb.22374. J Cell Biochem. 2010. PMID: 19899110 Free PMC article. Review.
-
Embryonic stem cell microRNAs: defining factors in induced pluripotent (iPS) and cancer (CSC) stem cells?Curr Stem Cell Res Ther. 2009 Sep;4(3):168-77. doi: 10.2174/157488809789057400. Curr Stem Cell Res Ther. 2009. PMID: 19492978 Review.
Cited by
-
DNA methylation and its basic function.Neuropsychopharmacology. 2013 Jan;38(1):23-38. doi: 10.1038/npp.2012.112. Epub 2012 Jul 11. Neuropsychopharmacology. 2013. PMID: 22781841 Free PMC article. Review.
-
Regulation of transposable elements by DNA modifications.Nat Rev Genet. 2019 Jul;20(7):417-431. doi: 10.1038/s41576-019-0106-6. Nat Rev Genet. 2019. PMID: 30867571 Review.
-
High expression of Endogenous Retroviruses from intrauterine life to adulthood in two mouse models of Autism Spectrum Disorders.Sci Rep. 2018 Jan 12;8(1):629. doi: 10.1038/s41598-017-19035-w. Sci Rep. 2018. PMID: 29330412 Free PMC article.
-
Vaccination with Ad5 vectors expands Ad5-specific CD8 T cells without altering memory phenotype or functionality.PLoS One. 2010 Dec 22;5(12):e14385. doi: 10.1371/journal.pone.0014385. PLoS One. 2010. PMID: 21203546 Free PMC article.
-
Retrotransposon-induced heterochromatin spreading in the mouse revealed by insertional polymorphisms.PLoS Genet. 2011 Sep;7(9):e1002301. doi: 10.1371/journal.pgen.1002301. Epub 2011 Sep 29. PLoS Genet. 2011. PMID: 21980304 Free PMC article.
References
-
- Bestor T. H. (2000) Hum. Mol. Genet. 9, 2395–2402 - PubMed
-
- Goll M. G., Bestor T. H. (2005) Annu. Rev. Biochem. 74, 481–514 - PubMed
-
- Robertson K. D., Wolffe A. P. (2000) Nat. Rev. Genet. 1, 11–19 - PubMed
-
- Jaenisch R., Bird A. (2003) Nat. Genet. 33, 245–254 - PubMed
-
- Klose R. J., Bird A. P. (2006) Trends Biochem. Sci. 31, 89–97 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

