The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci
- PMID: 20398922
- PMCID: PMC2879619
- DOI: 10.1016/j.cell.2010.03.010
The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci
Erratum in
- Cell. 2010 Oct 1;143(1):170
Abstract
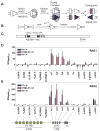
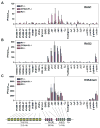
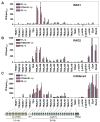
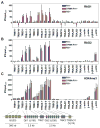
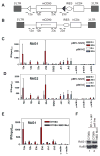
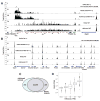
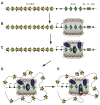
The critical initial step in V(D)J recombination, binding of RAG1 and RAG2 to recombination signal sequences flanking antigen receptor V, D, and J gene segments, has not previously been characterized in vivo. Here, we demonstrate that RAG protein binding occurs in a highly focal manner to a small region of active chromatin encompassing Ig kappa and Tcr alpha J gene segments and Igh and Tcr beta J and J-proximal D gene segments. Formation of these small RAG-bound regions, which we refer to as recombination centers, occurs in a developmental stage- and lineage-specific manner. Each RAG protein is independently capable of specific binding within recombination centers. While RAG1 binding was detected only at regions containing recombination signal sequences, RAG2 binds at thousands of sites in the genome containing histone 3 trimethylated at lysine 4. We propose that recombination centers coordinate V(D)J recombination by providing discrete sites within which gene segments are captured for recombination.
2010 Elsevier Inc. All rights reserved.
Figures
Comment in
-
Epigenetics drives RAGs to recombination riches.Cell. 2010 Apr 30;141(3):400-2. doi: 10.1016/j.cell.2010.04.014. Cell. 2010. PMID: 20434980
-
V(D)J recombination: RAG recombination centres.Nat Rev Immunol. 2010 Jun;10(6):383. doi: 10.1038/nri2789. Nat Rev Immunol. 2010. PMID: 20514673 No abstract available.
Similar articles
-
Partial reconstitution of V(D)J rearrangement and lymphocyte development in RAG-deficient mice expressing inducible, tetracycline-regulated RAG transgenes.Mol Immunol. 2004 Jan;40(11):813-29. doi: 10.1016/j.molimm.2003.09.009. Mol Immunol. 2004. PMID: 14687938
-
Noncore RAG1 regions promote Vβ rearrangements and αβ T cell development by overcoming inherent inefficiency of Vβ recombination signal sequences.J Immunol. 2014 Feb 15;192(4):1609-19. doi: 10.4049/jimmunol.1301599. Epub 2014 Jan 10. J Immunol. 2014. PMID: 24415779 Free PMC article.
-
V(D)J recombination: RAG proteins, repair factors, and regulation.Annu Rev Biochem. 2002;71:101-32. doi: 10.1146/annurev.biochem.71.090501.150203. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045092 Review.
-
Hypothesis: a biological role for germline transcription in the mechanism of V(D)J recombination--implications for initiation of allelic exclusion.Immunol Cell Biol. 2006 Aug;84(4):396-403. doi: 10.1111/j.1440-1711.2006.01437.x. Epub 2006 Apr 3. Immunol Cell Biol. 2006. PMID: 16594898
-
Activation of V(D)J recombination by RAG1 and RAG2.Trends Genet. 1992 Dec;8(12):413-6. doi: 10.1016/0168-9525(92)90323-v. Trends Genet. 1992. PMID: 1492366 Review.
Cited by
-
Setd1a regulates progenitor B-cell-to-precursor B-cell development through histone H3 lysine 4 trimethylation and Ig heavy-chain rearrangement.FASEB J. 2015 Apr;29(4):1505-15. doi: 10.1096/fj.14-263061. Epub 2014 Dec 30. FASEB J. 2015. PMID: 25550471 Free PMC article.
-
Cooperative recruitment of HMGB1 during V(D)J recombination through interactions with RAG1 and DNA.Nucleic Acids Res. 2013 Mar 1;41(5):3289-301. doi: 10.1093/nar/gks1461. Epub 2013 Jan 15. Nucleic Acids Res. 2013. PMID: 23325855 Free PMC article.
-
Clonal allelic predetermination of immunoglobulin-κ rearrangement.Nature. 2012 Oct 25;490(7421):561-5. doi: 10.1038/nature11496. Epub 2012 Sep 30. Nature. 2012. PMID: 23023124
-
Recombination may occur in the absence of transcription in the immunoglobulin heavy chain recombination centre.Nucleic Acids Res. 2020 Apr 17;48(7):3553-3566. doi: 10.1093/nar/gkaa108. Nucleic Acids Res. 2020. PMID: 32086526 Free PMC article.
-
CTCF-binding elements mediate control of V(D)J recombination.Nature. 2011 Sep 11;477(7365):424-30. doi: 10.1038/nature10495. Nature. 2011. PMID: 21909113 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Berg LJ, Pullen AM, Fazekas de St Groth B, Mathis D, Benoist C, Davis MM. Antigen/MHC-specific T cells are preferentially exported from the thymus in the presence of their MHC ligand. Cell. 1989;58:1035–1046. - PubMed
-
- Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. - PubMed
-
- Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova I, Sen R. Repeat organization and epigenetic regulation of the D-H-C-mu domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

