SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor
- PMID: 20388728
- PMCID: PMC2861189
- DOI: 10.1101/gad.1906510
SUMO-independent in vivo activity of a SUMO-targeted ubiquitin ligase toward a short-lived transcription factor
Abstract
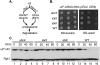
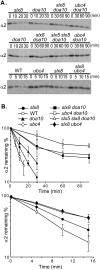
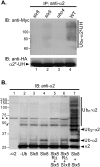
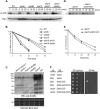
Many proteins are regulated by ubiquitin-dependent proteolysis. Substrate ubiquitylation can be stimulated by additional post-translational modifications, including small ubiquitin-like modifier (SUMO) conjugation. The recently discovered SUMO-targeted ubiquitin ligases (STUbLs) mediate the latter effect; however, no endogenous substrates of STUbLs that are degraded under normal conditions are known. From a targeted genomic screen, we now identify the yeast STUbL Slx5-Slx8, a heterodimeric RING protein complex, as a key ligase mediating degradation of the MATalpha2 (alpha2) repressor. The ubiquitin-conjugating enzyme Ubc4 was found in the same screen. Surprisingly, mutants with severe defects in SUMO-protein conjugation were not impaired for alpha2 turnover. Unmodified alpha2 also bound to and was ubiquitylated efficiently by Slx5-Slx8. Nevertheless, when we inactivated four SUMO-interacting motifs (SIMs) in Slx5 that together account for its noncovalent SUMO binding, both in vitro Slx5-Slx8-dependent ubiquitylation and in vivo degradation of alpha2 were inhibited. These data identify alpha2 as the first native substrate of the conserved STUbLs, and demonstrate that its STUbL-mediated ubiquitylation does not require SUMO. We suggest that alpha2, and presumably other proteins, have surface features that mimic SUMO, and therefore can directly recruit STUbLs without prior SUMO conjugation.
Figures

Similar articles
-
STUbL-mediated degradation of the transcription factor MATα2 requires degradation elements that coincide with corepressor binding sites.Mol Biol Cell. 2015 Oct 1;26(19):3401-12. doi: 10.1091/mbc.E15-06-0436. Epub 2015 Aug 5. Mol Biol Cell. 2015. PMID: 26246605 Free PMC article.
-
A SUMO-targeted ubiquitin ligase is involved in the degradation of the nuclear pool of the SUMO E3 ligase Siz1.Mol Biol Cell. 2014 Jan;25(1):1-16. doi: 10.1091/mbc.E13-05-0291. Epub 2013 Nov 6. Mol Biol Cell. 2014. PMID: 24196836 Free PMC article.
-
SUMO-targeted ubiquitin ligases in genome stability.EMBO J. 2007 Sep 19;26(18):4089-101. doi: 10.1038/sj.emboj.7601838. Epub 2007 Aug 30. EMBO J. 2007. PMID: 17762865 Free PMC article.
-
A SIM-ultaneous role for SUMO and ubiquitin.Trends Biochem Sci. 2008 May;33(5):201-8. doi: 10.1016/j.tibs.2008.02.001. Epub 2008 Apr 9. Trends Biochem Sci. 2008. PMID: 18403209 Review.
-
Nuclear organization in genome stability: SUMO connections.Cell Res. 2011 Mar;21(3):474-85. doi: 10.1038/cr.2011.31. Epub 2011 Feb 15. Cell Res. 2011. PMID: 21321608 Free PMC article. Review.
Cited by
-
SUMO-Targeted Ubiquitin Ligases (STUbLs) Reduce the Toxicity and Abnormal Transcriptional Activity Associated With a Mutant, Aggregation-Prone Fragment of Huntingtin.Front Genet. 2018 Sep 18;9:379. doi: 10.3389/fgene.2018.00379. eCollection 2018. Front Genet. 2018. PMID: 30279700 Free PMC article.
-
SUMO-Chain-Regulated Proteasomal Degradation Timing Exemplified in DNA Replication Initiation.Mol Cell. 2019 Nov 21;76(4):632-645.e6. doi: 10.1016/j.molcel.2019.08.003. Epub 2019 Sep 10. Mol Cell. 2019. PMID: 31519521 Free PMC article.
-
The mRNA export adaptor Yra1 contributes to DNA double-strand break repair through its C-box domain.PLoS One. 2019 Apr 5;14(4):e0206336. doi: 10.1371/journal.pone.0206336. eCollection 2019. PLoS One. 2019. PMID: 30951522 Free PMC article.
-
The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition.Nat Rev Mol Cell Biol. 2010 Dec;11(12):861-71. doi: 10.1038/nrm3011. Nat Rev Mol Cell Biol. 2010. PMID: 21102611 Free PMC article. Review.
-
DNA Damage Tolerance Pathway Choice Through Uls1 Modulation of Srs2 SUMOylation in Saccharomyces cerevisiae.Genetics. 2017 May;206(1):513-525. doi: 10.1534/genetics.116.196568. Epub 2017 Mar 24. Genetics. 2017. PMID: 28341648 Free PMC article.
References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K 1989. Current protocols in molecular biology Wiley, New York
-
- Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol 3: 24–29 - PubMed
-
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell 74: 357–369 - PubMed
-
- Deng M, Hochstrasser M 2006. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 443: 827–831 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
