SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism
- PMID: 20386712
- PMCID: PMC2851658
- DOI: 10.1371/journal.ppat.1000849
SARS-CoV pathogenesis is regulated by a STAT1 dependent but a type I, II and III interferon receptor independent mechanism
Abstract
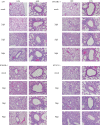
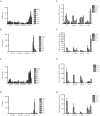
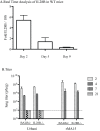
Severe acute respiratory syndrome coronavirus (SARS-CoV) infection often caused severe end stage lung disease and organizing phase diffuse alveolar damage, especially in the elderly. The virus-host interactions that governed development of these acute end stage lung diseases and death are unknown. To address this question, we evaluated the role of innate immune signaling in protection from human (Urbani) and a recombinant mouse adapted SARS-CoV, designated rMA15. In contrast to most models of viral pathogenesis, infection of type I, type II or type III interferon knockout mice (129 background) with either Urbani or MA15 viruses resulted in clinical disease outcomes, including transient weight loss, denuding bronchiolitis and alveolar inflammation and recovery, identical to that seen in infection of wildtype mice. This suggests that type I, II and III interferon signaling play minor roles in regulating SARS pathogenesis in mouse models. In contrast, infection of STAT1-/- mice resulted in severe disease, high virus titer, extensive pulmonary lesions and 100% mortality by day 9 and 30 post-infection with rMA15 or Urbani viruses, respectively. Non-lethal in BALB/c mice, Urbani SARS-CoV infection in STAT1-/- mice caused disseminated infection involving the liver, spleen and other tissues after day 9. These findings demonstrated that SARS-CoV pathogenesis is regulated by a STAT1 dependent but type I, II and III interferon receptor independent, mechanism. In contrast to a well documented role in innate immunity, we propose that STAT1 also protects mice via its role as an antagonist of unrestrained cell proliferation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
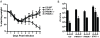

Similar articles
-
Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread.J Gen Virol. 2012 Dec;93(Pt 12):2601-2605. doi: 10.1099/vir.0.046284-0. Epub 2012 Sep 5. J Gen Virol. 2012. PMID: 22956738
-
Induction of alternatively activated macrophages enhances pathogenesis during severe acute respiratory syndrome coronavirus infection.J Virol. 2012 Dec;86(24):13334-49. doi: 10.1128/JVI.01689-12. Epub 2012 Sep 26. J Virol. 2012. PMID: 23015710 Free PMC article.
-
Transcriptomic analysis reveals a mechanism for a prefibrotic phenotype in STAT1 knockout mice during severe acute respiratory syndrome coronavirus infection.J Virol. 2010 Nov;84(21):11297-309. doi: 10.1128/JVI.01130-10. Epub 2010 Aug 11. J Virol. 2010. PMID: 20702617 Free PMC article.
-
SARS coronavirus and innate immunity.Virus Res. 2008 Apr;133(1):101-12. doi: 10.1016/j.virusres.2007.03.015. Epub 2007 Apr 23. Virus Res. 2008. PMID: 17451827 Free PMC article. Review.
-
Pathogenesis of severe acute respiratory syndrome.Curr Opin Immunol. 2005 Aug;17(4):404-10. doi: 10.1016/j.coi.2005.05.009. Curr Opin Immunol. 2005. PMID: 15950449 Free PMC article. Review.
Cited by
-
Type III interferons disrupt the lung epithelial barrier upon viral recognition.Science. 2020 Aug 7;369(6504):706-712. doi: 10.1126/science.abc3545. Epub 2020 Jun 11. Science. 2020. PMID: 32527925 Free PMC article.
-
Molecular pathology of emerging coronavirus infections.J Pathol. 2015 Jan;235(2):185-95. doi: 10.1002/path.4454. J Pathol. 2015. PMID: 25270030 Free PMC article. Review.
-
On Deep Landscape Exploration of COVID-19 Patients Cells and Severity Markers.Front Immunol. 2021 Sep 16;12:705646. doi: 10.3389/fimmu.2021.705646. eCollection 2021. Front Immunol. 2021. PMID: 34603282 Free PMC article.
-
Porcine Epidemic Diarrhea Virus Antagonizes Host IFN-λ-Mediated Responses by Tilting Transcription Factor STAT1 toward Acetylation over Phosphorylation To Block Its Activation.mBio. 2023 Jun 27;14(3):e0340822. doi: 10.1128/mbio.03408-22. Epub 2023 Apr 13. mBio. 2023. PMID: 37052505 Free PMC article.
-
Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling.mBio. 2010 Oct 26;1(5):e00206-10. doi: 10.1128/mBio.00206-10. mBio. 2010. PMID: 20978541 Free PMC article.
References
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. - PubMed
-
- Levy DE, Garcia-Sastre A. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 2001;12:143–156. - PubMed
-
- Seth RB, Sun L, Chen ZJ. Antiviral innate immunity pathways. Cell Res. 2006;16:141–147. - PubMed
-
- Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

