Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer
- PMID: 20385750
- PMCID: PMC2867283
- DOI: 10.1084/jem.20092437
Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer
Abstract
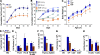
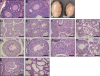
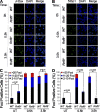
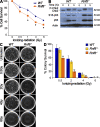
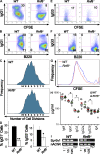
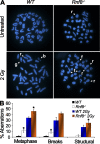
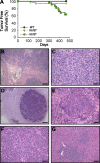
Signaling and repair of DNA double-strand breaks (DSBs) are critical for preventing immunodeficiency and cancer. These DNA breaks result from exogenous and endogenous DNA insults but are also programmed to occur during physiological processes such as meiosis and immunoglobulin heavy chain (IgH) class switch recombination (CSR). Recent studies reported that the E3 ligase RNF8 plays important roles in propagating DNA DSB signals and thereby facilitating the recruitment of various DNA damage response proteins, such as 53BP1 and BRCA1, to sites of damage. Using mouse models for Rnf8 mutation, we report that Rnf8 deficiency leads to impaired spermatogenesis and increased sensitivity to ionizing radiation both in vitro and in vivo. We also demonstrate the existence of alternative Rnf8-independent mechanisms that respond to irradiation and accounts for the partial recruitment of 53bp1 to sites of DNA damage in activated Rnf8(-/-) B cells. Remarkably, IgH CSR is impaired in a gene dose-dependent manner in Rnf8 mutant mice, revealing that these mice are immunodeficient. In addition, Rnf8(-/-) mice exhibit increased genomic instability and elevated risks for tumorigenesis indicating that Rnf8 is a novel tumor suppressor. These data unravel the in vivo pleiotropic effects of Rnf8.
Figures
Similar articles
-
Synergistic interaction of Rnf8 and p53 in the protection against genomic instability and tumorigenesis.PLoS Genet. 2013;9(1):e1003259. doi: 10.1371/journal.pgen.1003259. Epub 2013 Jan 31. PLoS Genet. 2013. PMID: 23382699 Free PMC article.
-
Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome.PLoS Genet. 2011 Apr;7(4):e1001381. doi: 10.1371/journal.pgen.1001381. Epub 2011 Apr 28. PLoS Genet. 2011. PMID: 21552324 Free PMC article.
-
Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8.J Exp Med. 2010 May 10;207(5):973-81. doi: 10.1084/jem.20092308. Epub 2010 Apr 12. J Exp Med. 2010. PMID: 20385748 Free PMC article.
-
RNF8-dependent histone ubiquitination during DNA damage response and spermatogenesis.Acta Biochim Biophys Sin (Shanghai). 2011 May;43(5):339-45. doi: 10.1093/abbs/gmr016. Epub 2011 Mar 28. Acta Biochim Biophys Sin (Shanghai). 2011. PMID: 21444325 Free PMC article. Review.
-
RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites.Cancer Lett. 2008 Nov 28;271(2):179-90. doi: 10.1016/j.canlet.2008.04.046. Epub 2008 Jun 11. Cancer Lett. 2008. PMID: 18550271 Free PMC article. Review.
Cited by
-
Rare Germline Variants in DNA Repair Genes Detected in BRCA-Negative Finnish Patients with Early-Onset Breast Cancer.Cancers (Basel). 2024 Aug 24;16(17):2955. doi: 10.3390/cancers16172955. Cancers (Basel). 2024. PMID: 39272813 Free PMC article.
-
Synergistic interaction of Rnf8 and p53 in the protection against genomic instability and tumorigenesis.PLoS Genet. 2013;9(1):e1003259. doi: 10.1371/journal.pgen.1003259. Epub 2013 Jan 31. PLoS Genet. 2013. PMID: 23382699 Free PMC article.
-
BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair.J Cell Biol. 2010 Oct 4;191(1):45-60. doi: 10.1083/jcb.201003034. J Cell Biol. 2010. PMID: 20921134 Free PMC article.
-
DNA double-strand break signaling and human disorders.Genome Integr. 2010 Nov 5;1(1):15. doi: 10.1186/2041-9414-1-15. Genome Integr. 2010. PMID: 21054854 Free PMC article.
-
Dimethyl Sulfoxide Attenuates Radiation-Induced Testicular Injury through Facilitating DNA Double-Strand Break Repair.Oxid Med Cell Longev. 2022 Jun 20;2022:9137812. doi: 10.1155/2022/9137812. eCollection 2022. Oxid Med Cell Longev. 2022. PMID: 35770047 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

