The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis
- PMID: 20385600
- PMCID: PMC2919705
- DOI: 10.1093/nar/gkq185
The C-terminus of Utp4, mutated in childhood cirrhosis, is essential for ribosome biogenesis
Abstract
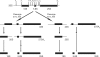
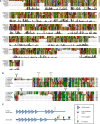
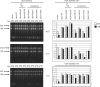
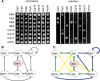
The small subunit (SSU) processome is a large ribonucleoprotein that is required for maturation of the 18S rRNA of the ribosome. Recently, a missense mutation in the C-terminus of an SSU processome protein, Utp4/Cirhin, was reported to cause North American Indian childhood cirrhosis (NAIC). In this study, we use Saccharomyces cerevisiae as a model to investigate the role of the NAIC mutation in ribosome biogenesis. While we find that the homologous NAIC mutation does not cause growth defects or aberrant ribosome biogenesis in yeast, we show that an intact C-terminus of Utp4 is required for cell growth and maturation of the 18S and 25S rRNAs. A protein-protein interaction map of the seven-protein t-Utp subcomplex of which Utp4 is a member shows that Utp8 interacts with the C-terminus of Utp4 and that this interaction is essential for assembly of the SSU processome and for the function of Utp4 in ribosome biogenesis. Furthermore, these results allow us to propose that NAIC may be caused by dysfunctional pre-ribosome assembly due to the loss of an interaction between the C-terminus of Utp4/Cirhin and another SSU processome protein.
Figures



Similar articles
-
NOL11, implicated in the pathogenesis of North American Indian childhood cirrhosis, is required for pre-rRNA transcription and processing.PLoS Genet. 2012;8(8):e1002892. doi: 10.1371/journal.pgen.1002892. Epub 2012 Aug 16. PLoS Genet. 2012. PMID: 22916032 Free PMC article.
-
p53-mediated biliary defects caused by knockdown of cirh1a, the zebrafish homolog of the gene responsible for North American Indian Childhood Cirrhosis.PLoS One. 2013 Oct 11;8(10):e77670. doi: 10.1371/journal.pone.0077670. eCollection 2013. PLoS One. 2013. PMID: 24147052 Free PMC article.
-
Human diseases of the SSU processome.Biochim Biophys Acta. 2014 Jun;1842(6):758-64. doi: 10.1016/j.bbadis.2013.11.004. Epub 2013 Nov 12. Biochim Biophys Acta. 2014. PMID: 24240090 Free PMC article. Review.
-
The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome.PLoS One. 2009 Dec 18;4(12):e8370. doi: 10.1371/journal.pone.0008370. PLoS One. 2009. PMID: 20019888 Free PMC article.
-
The small subunit processome in ribosome biogenesis—progress and prospects.Wiley Interdiscip Rev RNA. 2011 Jan-Feb;2(1):1-21. doi: 10.1002/wrna.57. Wiley Interdiscip Rev RNA. 2011. PMID: 21318072 Free PMC article. Review.
Cited by
-
Human nucleolar protein 7 (NOL7) is required for early pre-rRNA accumulation and pre-18S rRNA processing.RNA Biol. 2023 Jan;20(1):257-271. doi: 10.1080/15476286.2023.2217392. RNA Biol. 2023. PMID: 37246770 Free PMC article.
-
A Protein Microarray-Based Investigation of Cerebrospinal Fluid Reveals Distinct Autoantibody Signature in Low and High-Grade Gliomas.Front Oncol. 2020 Dec 22;10:543947. doi: 10.3389/fonc.2020.543947. eCollection 2020. Front Oncol. 2020. PMID: 33415070 Free PMC article.
-
Dysregulation of RNA polymerase I transcription during disease.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):342-60. doi: 10.1016/j.bbagrm.2012.10.014. Epub 2012 Nov 12. Biochim Biophys Acta. 2013. PMID: 23153826 Free PMC article. Review.
-
Ribosomopathies: mechanisms of disease.Clin Med Insights Blood Disord. 2014 Aug 14;7:7-16. doi: 10.4137/CMBD.S16952. eCollection 2014. Clin Med Insights Blood Disord. 2014. PMID: 25512719 Free PMC article. Review.
-
Cockayne syndrome group A and B proteins converge on transcription-linked resolution of non-B DNA.Proc Natl Acad Sci U S A. 2016 Nov 1;113(44):12502-12507. doi: 10.1073/pnas.1610198113. Epub 2016 Oct 18. Proc Natl Acad Sci U S A. 2016. PMID: 27791127 Free PMC article.
References
-
- Warner J. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. - PubMed
-
- Granneman S, Baserga S. Ribosome biogenesis: of knobs and RNA processing. Exp. Cell Res. 2004;296:43–50. - PubMed
-
- Dragon F, Gallagher J, Compagnone-Post P, Mitchell B, Porwancher K, Wehner K, Wormsley S, Settlage R, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. - PubMed
-
- Krogan N, Peng W, Cagney G, Robinson M, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards D, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

