Features and development of Coot
- PMID: 20383002
- PMCID: PMC2852313
- DOI: 10.1107/S0907444910007493
Features and development of Coot
Abstract
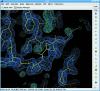
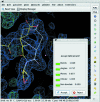
Coot is a molecular-graphics application for model building and validation of biological macromolecules. The program displays electron-density maps and atomic models and allows model manipulations such as idealization, real-space refinement, manual rotation/translation, rigid-body fitting, ligand search, solvation, mutations, rotamers and Ramachandran idealization. Furthermore, tools are provided for model validation as well as interfaces to external programs for refinement, validation and graphics. The software is designed to be easy to learn for novice users, which is achieved by ensuring that tools for common tasks are 'discoverable' through familiar user-interface elements (menus and toolbars) or by intuitive behaviour (mouse controls). Recent developments have focused on providing tools for expert users, with customisable key bindings, extensions and an extensive scripting interface. The software is under rapid development, but has already achieved very widespread use within the crystallographic community. The current state of the software is presented, with a description of the facilities available and of some of the underlying methods employed.
Figures

Similar articles
-
Current developments in Coot for macromolecular model building of Electron Cryo-microscopy and Crystallographic Data.Protein Sci. 2020 Apr;29(4):1069-1078. doi: 10.1002/pro.3791. Epub 2020 Mar 2. Protein Sci. 2020. PMID: 31730249 Free PMC article.
-
Handling ligands with Coot.Acta Crystallogr D Biol Crystallogr. 2012 Apr;68(Pt 4):425-30. doi: 10.1107/S0907444912000200. Epub 2012 Mar 16. Acta Crystallogr D Biol Crystallogr. 2012. PMID: 22505262 Free PMC article.
-
Coot: model-building tools for molecular graphics.Acta Crystallogr D Biol Crystallogr. 2004 Dec;60(Pt 12 Pt 1):2126-32. doi: 10.1107/S0907444904019158. Epub 2004 Nov 26. Acta Crystallogr D Biol Crystallogr. 2004. PMID: 15572765
-
Refinement of protein and nucleic acid structures.Methods Mol Biol. 1996;56:227-44. doi: 10.1385/0-89603-259-0:227. Methods Mol Biol. 1996. PMID: 8781248 Review. No abstract available.
-
Boxes of Model Building and Visualization.Methods Mol Biol. 2017;1607:491-548. doi: 10.1007/978-1-4939-7000-1_21. Methods Mol Biol. 2017. PMID: 28573587 Review.
Cited by
-
Structure and function of Mycobacterium tuberculosis EfpA as a lipid transporter and its inhibition by BRD-8000.3.Proc Natl Acad Sci U S A. 2024 Oct 29;121(44):e2412653121. doi: 10.1073/pnas.2412653121. Epub 2024 Oct 23. Proc Natl Acad Sci U S A. 2024. PMID: 39441632
-
Structural insights into the inhibition mechanism of fungal GWT1 by manogepix.Nat Commun. 2024 Oct 24;15(1):9194. doi: 10.1038/s41467-024-53512-x. Nat Commun. 2024. PMID: 39448635 Free PMC article.
-
Structures of the Varicella Zoster Virus Glycoprotein E and Epitope Mapping of Vaccine-Elicited Antibodies.Vaccines (Basel). 2024 Sep 27;12(10):1111. doi: 10.3390/vaccines12101111. Vaccines (Basel). 2024. PMID: 39460278 Free PMC article.
-
Piperidinols that show anti-tubercular activity as inhibitors of arylamine N-acetyltransferase: an essential enzyme for mycobacterial survival inside macrophages.PLoS One. 2012;7(12):e52790. doi: 10.1371/journal.pone.0052790. Epub 2012 Dec 28. PLoS One. 2012. PMID: 23285185 Free PMC article.
-
Conformational switches and redox properties of the colon cancer-associated human lectin ZG16.FEBS J. 2021 Nov;288(22):6465-6475. doi: 10.1111/febs.16044. Epub 2021 Jun 15. FEBS J. 2021. PMID: 34077620 Free PMC article.
References
-
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. - PubMed
-
- Bernstein, F. C., Koetzle, T. F., Williams, G. J. B., Meyer, E. F. Jr, Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol.112, 535–542. - PubMed
-
- Blanc, E., Roversi, P., Vonrhein, C., Flensburg, C., Lea, S. M. & Bricogne, G. (2004). Acta Cryst. D60, 2210–2221. - PubMed
-
- Blundell, T. L., Jhoti, H. & Abell, C. (2002). Nature Rev. Drug Discov.1, 45–54. - PubMed
-
- Brünger, A. T. (1992). X-PLOR v.3.1. A System for X-ray Crystallography and NMR. New Haven: Yale University Press.
Publication types
MeSH terms
Substances
Grants and funding
- BB/D522403/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/F020368/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM087721/GM/NIGMS NIH HHS/United States
- BB/D522403/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBF0202281/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

