Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway
- PMID: 20363318
- PMCID: PMC2893268
- DOI: 10.1016/j.cellsig.2010.03.017
Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway
Abstract
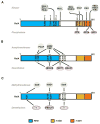
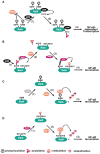
The eukaryotic transcription factor NF-kappaB regulates a wide range of host genes that control the inflammatory and immune responses, programmed cell death, cell proliferation and differentiation. The activation of NF-kappaB is tightly controlled both in the cytoplasm and in the nucleus. While the upstream cytoplasmic regulatory events for the activation of NF-kappaB are well studied, much less is known about the nuclear regulation of NF-kappaB. Emerging evidence suggests that NF-kappaB undergoes a variety of posttranslational modifications, and that these modifications play a key role in determining the duration and strength of NF-kappaB nuclear activity as well as its transcriptional output. Here we summarize the recent advances in our understanding of the posttranslational modifications of NF-kappaB, the interplay between the various modifications, and the physiological relevance of these modifications.
Published by Elsevier Inc.
Figures
Similar articles
-
Orf Virus ORF120 Protein Positively Regulates the NF-κB Pathway by Interacting with G3BP1.J Virol. 2021 Sep 9;95(19):e0015321. doi: 10.1128/JVI.00153-21. Epub 2021 Sep 9. J Virol. 2021. PMID: 34287041 Free PMC article.
-
Measuring NF-κB Phosphorylation and Acetylation.Methods Mol Biol. 2021;2366:3-17. doi: 10.1007/978-1-0716-1669-7_1. Methods Mol Biol. 2021. PMID: 34236629
-
Critical roles of IκBα and RelA phosphorylation in transitional oscillation in NF-κB signaling module.J Theor Biol. 2019 Feb 7;462:479-489. doi: 10.1016/j.jtbi.2018.11.023. Epub 2018 Nov 27. J Theor Biol. 2019. PMID: 30496749
-
Beyond IkappaBs: alternative regulation of NF-kappaB activity.FASEB J. 2007 Sep;21(11):2642-54. doi: 10.1096/fj.06-7615rev. Epub 2007 Apr 12. FASEB J. 2007. PMID: 17431096 Review.
-
Regulation of distinct biological activities of the NF-kappaB transcription factor complex by acetylation.J Mol Med (Berl). 2003 Sep;81(9):549-57. doi: 10.1007/s00109-003-0469-0. Epub 2003 Aug 15. J Mol Med (Berl). 2003. PMID: 12920522 Review.
Cited by
-
SNX25 regulates proinflammatory cytokine expression via the NF-κB signal in macrophages.PLoS One. 2021 Mar 1;16(3):e0247840. doi: 10.1371/journal.pone.0247840. eCollection 2021. PLoS One. 2021. PMID: 33647065 Free PMC article.
-
Retrospective Single Nucleotide Polymorphism Analysis of Host Resistance and Susceptibility to Ovine Johne's Disease Using Restored FFPE DNA.Int J Mol Sci. 2024 Jul 15;25(14):7748. doi: 10.3390/ijms25147748. Int J Mol Sci. 2024. PMID: 39062990 Free PMC article.
-
Multiplexing information flow through dynamic signalling systems.PLoS Comput Biol. 2020 Aug 3;16(8):e1008076. doi: 10.1371/journal.pcbi.1008076. eCollection 2020 Aug. PLoS Comput Biol. 2020. PMID: 32745094 Free PMC article.
-
Protein post-translational modifications in the regulation of cancer hallmarks.Cancer Gene Ther. 2023 Apr;30(4):529-547. doi: 10.1038/s41417-022-00464-3. Epub 2022 Apr 7. Cancer Gene Ther. 2023. PMID: 35393571 Review.
-
A direct nuclear magnetic resonance method to investigate lysine acetylation of intrinsically disordered proteins.Front Mol Biosci. 2023 Jan 6;9:1074743. doi: 10.3389/fmolb.2022.1074743. eCollection 2022. Front Mol Biosci. 2023. PMID: 36685286 Free PMC article.
References
-
- Baldwin AS., Jr The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. - PubMed
-
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. - PubMed
-
- Karin M. How NF-kappaB is activated: the role of the IkappaB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. - PubMed
-
- Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol. 2000;18:621–663. - PubMed
-
- Ghosh S, Karin M. Missing pieces in the NF-kappaB puzzle. Cell. 2002;109(Suppl):S81–S96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

