Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes
- PMID: 20357091
- PMCID: PMC2876602
- DOI: 10.1128/JVI.02200-09
Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes
Abstract
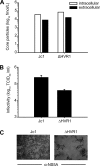
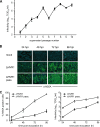
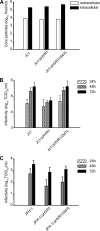
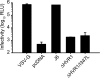
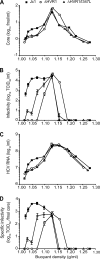
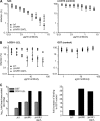
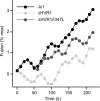
The variability of the hepatitis C virus (HCV), which likely contributes to immune escape, is most pronounced in hypervariable region 1 (HVR1) of viral envelope protein 2. This domain is the target for neutralizing antibodies, and its deletion attenuates replication in vivo. Here we characterized the relevance of HVR1 for virus replication in vitro using cell culture-derived HCV. We show that HVR1 is dispensable for RNA replication. However, viruses lacking HVR1 (Delta HVR1) are less infectious, and separation by density gradients revealed that the population of Delta HVR1 virions comprises fewer particles with low density. Strikingly, Delta HVR1 particles with intermediate density (1.12 g/ml) are as infectious as wild-type virions, while those with low density (1.02 to 1.08 g/ml) are poorly infectious, despite quantities of RNA and core similar to those in wild-type particles. Moreover, Delta HVR1 particles exhibited impaired fusion, a defect that was partially restored by an E1 mutation (I347L), which also rescues infectivity and which was selected during long-term culture. Finally, Delta HVR1 particles were no longer neutralized by SR-B1-specific immunoglobulins but were more prone to neutralization and precipitation by soluble CD81, E2-specific monoclonal antibodies, and patient sera. These results suggest that HVR1 influences the biophysical properties of released viruses and that this domain is particularly important for infectivity of low-density particles. Moreover, they indicate that HVR1 obstructs the viral CD81 binding site and conserved neutralizing epitopes. These functions likely optimize virus replication, facilitate immune escape, and thus foster establishment and maintenance of a chronic infection.
Figures

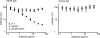

Similar articles
-
Functional and immunogenic characterization of diverse HCV glycoprotein E2 variants.J Hepatol. 2019 Apr;70(4):593-602. doi: 10.1016/j.jhep.2018.11.003. Epub 2018 Nov 13. J Hepatol. 2019. PMID: 30439392
-
Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity.J Virol. 2009 Jun;83(12):6149-60. doi: 10.1128/JVI.00248-09. Epub 2009 Mar 25. J Virol. 2009. PMID: 19321602 Free PMC article.
-
Hypervariable Region 1 in Envelope Protein 2 of Hepatitis C Virus: A Linchpin in Neutralizing Antibody Evasion and Viral Entry.Front Immunol. 2018 Sep 27;9:2146. doi: 10.3389/fimmu.2018.02146. eCollection 2018. Front Immunol. 2018. PMID: 30319614 Free PMC article. Review.
-
Role of hypervariable region 1 for the interplay of hepatitis C virus with entry factors and lipoproteins.J Virol. 2014 Nov;88(21):12644-55. doi: 10.1128/JVI.01145-14. Epub 2014 Aug 20. J Virol. 2014. PMID: 25142595 Free PMC article.
-
Virus-neutralizing antibodies to hepatitis C virus.J Viral Hepat. 2013 Jun;20(6):369-76. doi: 10.1111/jvh.12094. Epub 2013 Apr 4. J Viral Hepat. 2013. PMID: 23647953 Review.
Cited by
-
Induction of cross-neutralizing antibodies by a permuted hepatitis C virus glycoprotein nanoparticle vaccine candidate.Nat Commun. 2022 Nov 25;13(1):7271. doi: 10.1038/s41467-022-34961-8. Nat Commun. 2022. PMID: 36434005 Free PMC article.
-
Identification of transferrin receptor 1 as a hepatitis C virus entry factor.Proc Natl Acad Sci U S A. 2013 Jun 25;110(26):10777-82. doi: 10.1073/pnas.1301764110. Epub 2013 Jun 10. Proc Natl Acad Sci U S A. 2013. PMID: 23754414 Free PMC article.
-
Three different functional microdomains in the hepatitis C virus hypervariable region 1 (HVR1) mediate entry and immune evasion.J Biol Chem. 2012 Oct 12;287(42):35631-35645. doi: 10.1074/jbc.M112.382341. Epub 2012 Aug 27. J Biol Chem. 2012. PMID: 22927442 Free PMC article.
-
Hepatitis C Virus Vaccine: Challenges and Prospects.Vaccines (Basel). 2020 Feb 17;8(1):90. doi: 10.3390/vaccines8010090. Vaccines (Basel). 2020. PMID: 32079254 Free PMC article. Review.
-
Oligonucleotide-Lipid Conjugates Forming G-Quadruplex Structures Are Potent and Pangenotypic Hepatitis C Virus Entry Inhibitors In Vitro and Ex Vivo.Antimicrob Agents Chemother. 2017 Apr 24;61(5):e02354-16. doi: 10.1128/AAC.02354-16. Print 2017 May. Antimicrob Agents Chemother. 2017. PMID: 28193659 Free PMC article.
References
-
- Bartosch, B., G. Verney, M. Dreux, P. Donot, Y. Morice, F. Penin, J. M. Pawlotsky, D. Lavillette, and F. L. Cosset. 2005. An interplay between hypervariable region 1 of the hepatitis C virus E2 glycoprotein, the scavenger receptor BI, and high-density lipoprotein promotes both enhancement of infection and protection against neutralizing antibodies. J. Virol. 79:8217-8229. - PMC - PubMed
-
- Bartosch, B., A. Vitelli, C. Granier, C. Goujon, J. Dubuisson, S. Pascale, E. Scarselli, R. Cortese, A. Nicosia, and F. L. Cosset. 2003. Cell entry of hepatitis C virus requires a set of co-receptors that include the CD81 tetraspanin and the SR-B1 scavenger receptor. J. Biol. Chem. 278:41624-41630. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

