Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors
- PMID: 20357002
- PMCID: PMC2877642
- DOI: 10.1091/mbc.e10-01-0016
Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors
Abstract
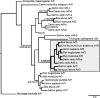
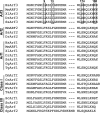
It is widely assumed that class I and II Arfs function interchangeably throughout the Golgi complex. However, we report here that in vivo, Arf3 displays several unexpected properties. Unlike other Golgi-localized Arfs, Arf3 associates selectively with membranes of the trans-Golgi network (TGN) in a manner that is both temperature-sensitive and uniquely dependent on guanine nucleotide exchange factors of the BIGs family. For example, BIGs knockdown redistributed Arf3 but not Arf1 from Golgi membranes. Furthermore, shifting temperature to 20 degrees C, a temperature known to block cargo in the TGN, selectively redistributed Arf3 from Golgi membranes. Arf3 redistribution occurred slowly, suggesting it resulted from a change in membrane composition. Arf3 knockdown and overexpression experiments suggest that redistribution is not responsible for the 20 degrees C block. To investigate in more detail the mechanism for Arf3 recruitment and temperature-dependent release, we characterized several mutant forms of Arf3. This analysis demonstrated that those properties are readily separated and depend on pairs of residues present at opposite ends of the protein. Furthermore, phylogenetic analysis established that all four critical residues were absolutely conserved and unique to Arf3. These results suggest that Arf3 plays a unique function at the TGN that likely involves recruitment by a specific receptor.
Figures


Similar articles
-
Distinct functions for Arf guanine nucleotide exchange factors at the Golgi complex: GBF1 and BIGs are required for assembly and maintenance of the Golgi stack and trans-Golgi network, respectively.Mol Biol Cell. 2008 Feb;19(2):523-35. doi: 10.1091/mbc.e07-04-0394. Epub 2007 Nov 14. Mol Biol Cell. 2008. PMID: 18003980 Free PMC article.
-
The Sec7 guanine nucleotide exchange factor GBF1 regulates membrane recruitment of BIG1 and BIG2 guanine nucleotide exchange factors to the trans-Golgi network (TGN).J Biol Chem. 2013 Apr 19;288(16):11532-45. doi: 10.1074/jbc.M112.438481. Epub 2013 Feb 5. J Biol Chem. 2013. PMID: 23386609 Free PMC article.
-
Dominant-negative mutant of BIG2, an ARF-guanine nucleotide exchange factor, specifically affects membrane trafficking from the trans-Golgi network through inhibiting membrane association of AP-1 and GGA coat proteins.Biochem Biophys Res Commun. 2002 Jun 7;294(2):254-60. doi: 10.1016/S0006-291X(02)00456-4. Biochem Biophys Res Commun. 2002. PMID: 12051703
-
Class II Arfs require a brefeldin-A-sensitive factor for Golgi association.Biochem Biophys Res Commun. 2020 Sep 10;530(1):301-306. doi: 10.1016/j.bbrc.2020.07.001. Epub 2020 Aug 6. Biochem Biophys Res Commun. 2020. PMID: 32828303
-
Class I Arfs (Arf1 and Arf3) and Arf6 are localized to the Flemming body and play important roles in cytokinesis.J Biochem. 2016 Feb;159(2):201-8. doi: 10.1093/jb/mvv088. Epub 2015 Sep 1. J Biochem. 2016. PMID: 26330566 Free PMC article.
Cited by
-
Kinetics of Arf1 inactivation regulates Golgi organisation and function in non-adherent fibroblasts.Biol Open. 2023 Apr 15;12(4):bio059669. doi: 10.1242/bio.059669. Epub 2023 May 4. Biol Open. 2023. PMID: 36946871 Free PMC article.
-
Dominant ARF3 variants disrupt Golgi integrity and cause a neurodevelopmental disorder recapitulated in zebrafish.Nat Commun. 2022 Nov 11;13(1):6841. doi: 10.1038/s41467-022-34354-x. Nat Commun. 2022. PMID: 36369169 Free PMC article.
-
Class I ADP-ribosylation factors are involved in enterovirus 71 replication.PLoS One. 2014 Jun 9;9(6):e99768. doi: 10.1371/journal.pone.0099768. eCollection 2014. PLoS One. 2014. PMID: 24911624 Free PMC article.
-
Ancient complement and lineage-specific evolution of the Sec7 ARF GEF proteins in eukaryotes.Mol Biol Cell. 2019 Jul 15;30(15):1846-1863. doi: 10.1091/mbc.E19-01-0073. Epub 2019 May 29. Mol Biol Cell. 2019. PMID: 31141460 Free PMC article.
-
Arf GTPases Are Required for the Establishment of the Pre-Assembly Compartment in the Early Phase of Cytomegalovirus Infection.Life (Basel). 2021 Aug 23;11(8):867. doi: 10.3390/life11080867. Life (Basel). 2021. PMID: 34440611 Free PMC article.
References
-
- Abascal F., Zardoya R., Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Antonny B., Beraud-Dufour S., Chardin P., Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. - PubMed
-
- Bard F., Malhotra V. The formation of TGN-to-plasma-membrane transport carriers. Annu. Rev. Cell Dev. Biol. 2006;22:439–455. - PubMed
-
- Berger S. J., Claude A., Melançon P. Analysis of recombitant human ADP-ribosylation factors by reversed phase HPLC and Electrospray MS. Analyt. Biochem. 1998;264:53–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

