Comparative analysis of chromatin binding by Sex Comb on Midleg (SCM) and other polycomb group repressors at a Drosophila Hox gene
- PMID: 20351181
- PMCID: PMC2876513
- DOI: 10.1128/MCB.01451-09
Comparative analysis of chromatin binding by Sex Comb on Midleg (SCM) and other polycomb group repressors at a Drosophila Hox gene
Erratum in
-
Correction for Wang et al., "Comparative Analysis of Chromatin Binding by Sex Comb on Midleg (SCM) and Other Polycomb Group Repressors at a Drosophila Hox Gene".Mol Cell Biol. 2017 Jul 14;37(15):e00148-17. doi: 10.1128/MCB.00148-17. Print 2017 Aug 1. Mol Cell Biol. 2017. PMID: 28710118 Free PMC article. No abstract available.
Abstract
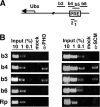
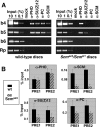
Sex Comb on Midleg (SCM) is a transcriptional repressor in the Polycomb group (PcG), but its molecular role in PcG silencing is not known. Although SCM can interact with Polycomb repressive complex 1 (PRC1) in vitro, biochemical studies have indicated that SCM is not a core constituent of PRC1 or PRC2. Nevertheless, SCM is just as critical for Drosophila Hox gene silencing as canonical subunits of these well-characterized PcG complexes. To address functional relationships between SCM and other PcG components, we have performed chromatin immunoprecipitation studies using cultured Drosophila Schneider line 2 (S2) cells and larval imaginal discs. We find that SCM associates with a Polycomb response element (PRE) upstream of the Ubx gene which also binds PRC1, PRC2, and the DNA-binding PcG protein Pleiohomeotic (PHO). However, SCM is retained at this Ubx PRE despite genetic disruption or knockdown of PHO, PRC1, or PRC2, suggesting that SCM chromatin targeting does not require prior association of these other PcG components. Chromatin immunoprecipitations (IPs) to test the consequences of SCM genetic disruption or knockdown revealed that PHO association is unaffected, but reduced levels of PRE-bound PRC2 and PRC1 were observed. We discuss these results in light of current models for recruitment of PcG complexes to chromatin targets.
Figures



Similar articles
-
Sex comb on midleg (Scm) is a functional link between PcG-repressive complexes in Drosophila.Genes Dev. 2015 Jun 1;29(11):1136-50. doi: 10.1101/gad.260562.115. Genes Dev. 2015. PMID: 26063573 Free PMC article.
-
Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression.Genetics. 2004 Jul;167(3):1225-39. doi: 10.1534/genetics.104.027474. Genetics. 2004. PMID: 15280237 Free PMC article.
-
The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing.Development. 2003 Jan;130(2):285-94. doi: 10.1242/dev.00204. Development. 2003. PMID: 12466196
-
Chromatin regulation: how complex does it get?Epigenetics. 2014 Nov;9(11):1485-95. doi: 10.4161/15592294.2014.971580. Epigenetics. 2014. PMID: 25482055 Free PMC article. Review.
-
Polycomb-mediated gene silencing in Arabidopsis thaliana.Mol Cells. 2014 Dec 31;37(12):841-50. doi: 10.14348/molcells.2014.0249. Epub 2014 Nov 20. Mol Cells. 2014. PMID: 25410906 Free PMC article. Review.
Cited by
-
Elements of the polycomb repressor SU(Z)12 needed for histone H3-K27 methylation, the interface with E(Z), and in vivo function.Mol Cell Biol. 2013 Dec;33(24):4844-56. doi: 10.1128/MCB.00307-13. Epub 2013 Oct 7. Mol Cell Biol. 2013. PMID: 24100017 Free PMC article.
-
Spps, a Drosophila Sp1/KLF family member, binds to PREs and is required for PRE activity late in development.Development. 2010 Aug 1;137(15):2597-602. doi: 10.1242/dev.047761. Development. 2010. PMID: 20627963 Free PMC article.
-
Identification and characterization of Polycomb group genes in the silkworm, Bombyx mori.Mol Biol Rep. 2012 May;39(5):5575-88. doi: 10.1007/s11033-011-1362-5. Epub 2011 Dec 21. Mol Biol Rep. 2012. PMID: 22187347
-
Sex Comb on Midleg Like-2 Accelerates Hepatocellular Carcinoma Cell Proliferation and Metastasis by Activating Wnt/β-Catenin/EMT Signaling.Yonsei Med J. 2021 Dec;62(12):1073-1082. doi: 10.3349/ymj.2021.62.12.1073. Yonsei Med J. 2021. PMID: 34816637 Free PMC article.
-
Recruitment of polycomb complexes: a role for SCM.Mol Cell Biol. 2010 Jun;30(11):2581-3. doi: 10.1128/MCB.00231-10. Epub 2010 Mar 29. Mol Cell Biol. 2010. PMID: 20351178 Free PMC article. No abstract available.
References
-
- Beuchle, D., G. Struhl, and J. Muller. 2001. Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128:993-1004. - PubMed
-
- Bornemann, D., E. Miller, and J. Simon. 1996. The Drosophila Polycomb group gene Sex comb on midleg (Scm) encodes a zinc finger protein with similarity to polyhomeotic protein. Development 122:1621-1630. - PubMed
-
- Boyer, L. A., K. Plath, J. Zeitlinger, T. Brambrink, L. A. Medeiros, T. I. Lee, S. S. Levine, M. Wernig, A. Tajonar, M. K. Ray, G. W. Bell, A. P. Otte, M. Vidal, D. K. Gifford, R. A. Young, and R. Jaenisch. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349-353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
