Differential regulation of microRNA stability
- PMID: 20348442
- PMCID: PMC2856875
- DOI: 10.1261/rna.1851510
Differential regulation of microRNA stability
Abstract
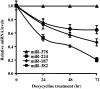
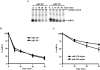
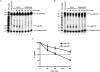
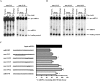
MicroRNAs (miRNAs) are endogenous single-stranded RNA molecules of about 21 nucleotides in length that are fundamental post-transcriptional regulators of gene expression. Although the transcriptional and processing events involved in the generation of miRNAs have been extensively studied, very little is known pertaining to components that regulate the stability of individual miRNAs. All RNAs have distinct inherent half-lives that dictate their level of accumulation and miRNAs would be expected to follow a similar principle. Here we demonstrate that although most miRNA appear to be stable, like mRNAs, miRNAs possess differential stability in human cells. In particular, we found that miR-382, a miRNA that contributes to HIV-1 provirus latency, is unstable in cells. To determine the region of miR-382 responsible for its rapid decay, we developed a cell-free system that recapitulated the observed cell-based-regulated miR-382 turnover. The system utilizes in vitro-processed mature miRNA derived from pre-miRNA and follows the decay of the processed miRNA. Using this system, we demonstrate that instability of miR-382 is driven by sequences outside its seed region and required the 3' terminal seven nucleotides where mutations in this region increased the stability of the RNA. Moreover, the exosome 3'-5' exoribonuclease complex was identified as the primary nuclease involved in miR-382 decay with a more modest contribution by the Xrn1 and no detectable contribution by Xrn2. These studies provide evidence for an miRNA element essential for rapid miRNA decay and implicate the exosome in this process. The development of a biochemically amendable system to analyze the mechanism of differential miRNA stability provides an important step in efforts to regulate gene expression by modulating miRNA stability.
Figures
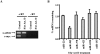
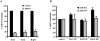
Similar articles
-
Triplex-forming MicroRNAs form stable complexes with HIV-1 provirus and inhibit its replication.Appl Immunohistochem Mol Morphol. 2010 Dec;18(6):532-45. doi: 10.1097/PAI.0b013e3181e1ef6a. Appl Immunohistochem Mol Morphol. 2010. PMID: 20502318
-
Analysis of microRNA turnover in mammalian cells following Dicer1 ablation.Nucleic Acids Res. 2011 Jul;39(13):5692-703. doi: 10.1093/nar/gkr148. Epub 2011 Mar 29. Nucleic Acids Res. 2011. PMID: 21447562 Free PMC article.
-
Role of pri-miRNA tertiary structure in miR-17~92 miRNA biogenesis.RNA Biol. 2011 Nov-Dec;8(6):1105-14. doi: 10.4161/rna.8.6.17410. Epub 2011 Nov 1. RNA Biol. 2011. PMID: 21955497
-
MicroRNA biogenesis: isolation and characterization of the microprocessor complex.Methods Mol Biol. 2006;342:33-47. doi: 10.1385/1-59745-123-1:33. Methods Mol Biol. 2006. PMID: 16957365 Review.
-
MicroRNA Expression: Protein Participants in MicroRNA Regulation.Methods Mol Biol. 2017;1617:27-37. doi: 10.1007/978-1-4939-7046-9_2. Methods Mol Biol. 2017. PMID: 28540674 Review.
Cited by
-
Defining a new role of GW182 in maintaining miRNA stability.EMBO Rep. 2012 Dec;13(12):1102-8. doi: 10.1038/embor.2012.160. Epub 2012 Oct 23. EMBO Rep. 2012. PMID: 23090477 Free PMC article.
-
Quantitative Analysis of MicroRNAs in Vaccinia virus Infection Reveals Diversity in Their Susceptibility to Modification and Suppression.PLoS One. 2015 Jul 10;10(7):e0131787. doi: 10.1371/journal.pone.0131787. eCollection 2015. PLoS One. 2015. PMID: 26161560 Free PMC article.
-
Roles for microRNAs in conferring robustness to biological processes.Cell. 2012 Apr 27;149(3):515-24. doi: 10.1016/j.cell.2012.04.005. Cell. 2012. PMID: 22541426 Free PMC article. Review.
-
MicroRNA degradation and turnover: regulating the regulators.Wiley Interdiscip Rev RNA. 2012 Jul-Aug;3(4):593-600. doi: 10.1002/wrna.1114. Epub 2012 Mar 28. Wiley Interdiscip Rev RNA. 2012. PMID: 22461385 Free PMC article. Review.
-
Target RNAs Strike Back on MicroRNAs.Front Genet. 2018 Oct 2;9:435. doi: 10.3389/fgene.2018.00435. eCollection 2018. Front Genet. 2018. PMID: 30333855 Free PMC article. Review.
References
-
- Amberg DC, Goldstein AL, Cole CN 1992. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev 6: 1173–1189 - PubMed
-
- Bentwich I 2005. Prediction and validation of microRNAs and their targets. FEBS Lett 579: 5904–5910 - PubMed
-
- Chatterjee S, Grosshans H 2009. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461: 546–549 - PubMed
-
- Coller J, Parker R 2004. Eukaryotic mRNA Decapping. Annu Rev Biochem 73: 861–890 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
