Origins of specificity in protein-DNA recognition
- PMID: 20334529
- PMCID: PMC3285485
- DOI: 10.1146/annurev-biochem-060408-091030
Origins of specificity in protein-DNA recognition
Abstract
Specific interactions between proteins and DNA are fundamental to many biological processes. In this review, we provide a revised view of protein-DNA interactions that emphasizes the importance of the three-dimensional structures of both macromolecules. We divide protein-DNA interactions into two categories: those when the protein recognizes the unique chemical signatures of the DNA bases (base readout) and those when the protein recognizes a sequence-dependent DNA shape (shape readout). We further divide base readout into those interactions that occur in the major groove from those that occur in the minor groove. Analogously, the readout of the DNA shape is subdivided into global shape recognition (for example, when the DNA helix exhibits an overall bend) and local shape recognition (for example, when a base pair step is kinked or a region of the minor groove is narrow). Based on the >1500 structures of protein-DNA complexes now available in the Protein Data Bank, we argue that individual DNA-binding proteins combine multiple readout mechanisms to achieve DNA-binding specificity. Specificity that distinguishes between families frequently involves base readout in the major groove, whereas shape readout is often exploited for higher resolution specificity, to distinguish between members within the same DNA-binding protein family.
Figures

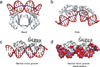
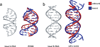

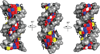

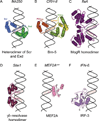
Similar articles
-
P22 c2 repressor-operator complex: mechanisms of direct and indirect readout.Biochemistry. 2008 Feb 26;47(8):2325-38. doi: 10.1021/bi701826f. Epub 2008 Feb 1. Biochemistry. 2008. PMID: 18237194
-
Indirect readout of DNA sequence at the primary-kink site in the CAP-DNA complex: DNA binding specificity based on energetics of DNA kinking.J Mol Biol. 2001 Nov 16;314(1):63-74. doi: 10.1006/jmbi.2001.5089. J Mol Biol. 2001. PMID: 11724532
-
The role of DNA shape in protein-DNA recognition.Nature. 2009 Oct 29;461(7268):1248-53. doi: 10.1038/nature08473. Nature. 2009. PMID: 19865164 Free PMC article.
-
DNA curving and bending in protein-DNA recognition.Mol Microbiol. 1992 Sep;6(18):2549-55. doi: 10.1111/j.1365-2958.1992.tb01431.x. Mol Microbiol. 1992. PMID: 1333034 Review.
-
Making the bend: DNA tertiary structure and protein-DNA interactions.Int J Mol Sci. 2014 Jul 14;15(7):12335-63. doi: 10.3390/ijms150712335. Int J Mol Sci. 2014. PMID: 25026169 Free PMC article. Review.
Cited by
-
Probing allostery through DNA.Science. 2013 Feb 15;339(6121):816-9. doi: 10.1126/science.1229223. Science. 2013. PMID: 23413354 Free PMC article.
-
Role of Shape Deformation of DNA-Binding Sites in Regulating the Efficiency and Specificity in Their Recognition by DNA-Binding Proteins.JACS Au. 2024 Jun 18;4(7):2640-2655. doi: 10.1021/jacsau.4c00393. eCollection 2024 Jul 22. JACS Au. 2024. PMID: 39055163 Free PMC article.
-
A robust assay to measure DNA topology-dependent protein binding affinity.Nucleic Acids Res. 2015 Apr 20;43(7):e43. doi: 10.1093/nar/gku1381. Epub 2014 Dec 30. Nucleic Acids Res. 2015. PMID: 25552413 Free PMC article.
-
Crosstalk between the HpArsRS two-component system and HpNikR is necessary for maximal activation of urease transcription.Front Microbiol. 2015 Jun 12;6:558. doi: 10.3389/fmicb.2015.00558. eCollection 2015. Front Microbiol. 2015. PMID: 26124751 Free PMC article.
-
Quantitative modeling of gene expression using DNA shape features of binding sites.Nucleic Acids Res. 2016 Jul 27;44(13):e120. doi: 10.1093/nar/gkw446. Epub 2016 Jun 1. Nucleic Acids Res. 2016. PMID: 27257066 Free PMC article.
References
-
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

