Interchromosomal association and gene regulation in trans
- PMID: 20236724
- PMCID: PMC2865229
- DOI: 10.1016/j.tig.2010.01.007
Interchromosomal association and gene regulation in trans
Abstract
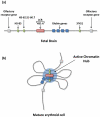
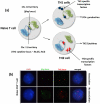
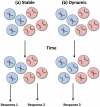
The nucleus is an ordered three-dimensional entity, and organization of the genome within the nuclear space might have implications for orchestrating gene expression. Recent technological developments have revealed that chromatin is folded into loops bringing distal regulatory elements into intimate contact with the genes that they regulate. Such intrachromosomal contacts appear to be a general mechanism of enhancer-promoter communication in cis. Tantalizing evidence is emerging that regulatory elements might have the capacity to act in trans to regulate genes on other chromosomes. However, unequivocal data required to prove that interchromosomal gene regulation truly represents another level of control within the nucleus is lacking, and this concept remains highly contentious. Such controversy emphasizes that our current understanding of the mechanisms that govern gene expression are far from complete.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Interchromosomal associations between alternatively expressed loci.Nature. 2005 Jun 2;435(7042):637-45. doi: 10.1038/nature03574. Epub 2005 May 8. Nature. 2005. PMID: 15880101
-
LHX2- and LDB1-mediated trans interactions regulate olfactory receptor choice.Nature. 2019 Jan;565(7740):448-453. doi: 10.1038/s41586-018-0845-0. Epub 2019 Jan 9. Nature. 2019. PMID: 30626972 Free PMC article.
-
Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D.Mol Cell. 2019 Jun 20;74(6):1110-1122. doi: 10.1016/j.molcel.2019.05.032. Mol Cell. 2019. PMID: 31226276 Review.
-
The interdependence of gene-regulatory elements and the 3D genome.J Cell Biol. 2019 Jan 7;218(1):12-26. doi: 10.1083/jcb.201809040. Epub 2018 Nov 15. J Cell Biol. 2019. PMID: 30442643 Free PMC article. Review.
-
Spatial organization of gene expression: the active chromatin hub.Chromosome Res. 2003;11(5):447-59. doi: 10.1023/a:1024922626726. Chromosome Res. 2003. PMID: 12971721 Review.
Cited by
-
Mechanism of KIT gene regulation by GATA1 lacking the N-terminal domain in Down syndrome-related myeloid disorders.Sci Rep. 2022 Nov 29;12(1):20587. doi: 10.1038/s41598-022-25046-z. Sci Rep. 2022. PMID: 36447001 Free PMC article.
-
Centromeric motion facilitates the mobility of interphase genomic regions in fission yeast.J Cell Sci. 2013 Nov 15;126(Pt 22):5271-83. doi: 10.1242/jcs.133678. Epub 2013 Aug 28. J Cell Sci. 2013. PMID: 23986481 Free PMC article.
-
The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading.PLoS Genet. 2013 Oct;9(10):e1003883. doi: 10.1371/journal.pgen.1003883. Epub 2013 Oct 31. PLoS Genet. 2013. PMID: 24204298 Free PMC article.
-
A new approach of gene co-expression network inference reveals significant biological processes involved in porcine muscle development in late gestation.Sci Rep. 2018 Jul 5;8(1):10150. doi: 10.1038/s41598-018-28173-8. Sci Rep. 2018. PMID: 29977047 Free PMC article.
-
A p53 enhancer region regulates target genes through chromatin conformations in cis and in trans.Genes Dev. 2013 Nov 15;27(22):2433-8. doi: 10.1101/gad.225565.113. Genes Dev. 2013. PMID: 24240233 Free PMC article.
References
-
- Cremer T, et al. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum Genet. 1982;60:46–56. - PubMed
-
- Tanabe H, et al. Inter- and intra-specific gene-density-correlated radial chromosome territory arrangements are conserved in Old World monkeys. Cytogenet Genome Res. 2005;108:255–261. - PubMed
-
- Mora L, et al. Chromosome territory positioning of conserved homologous chromosomes in different primate species. Chromosoma. 2006;115:367–375. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

