CXC chemokine receptor 4 is a cell surface receptor for extracellular ubiquitin
- PMID: 20228059
- PMCID: PMC2865327
- DOI: 10.1074/jbc.M110.103408
CXC chemokine receptor 4 is a cell surface receptor for extracellular ubiquitin
Abstract
Ubiquitin is one of the most highly conserved proteins in eukaryotes and plays major biological roles as a post-translational protein modifier. Ubiquitin is also a natural constituent of plasma, and several lines of evidence suggest that extracellular ubiquitin is an immune modulator with anti-inflammatory properties. In addition, ubiquitin treatment has been shown to limit inflammation and reduce organ injury in various disease models and species in vivo. However, its mechanism of action is unknown. Here we show that extracellular ubiquitin is a natural CXC chemokine receptor 4 (CXCR4 and CD184) agonist. Extracellular ubiquitin promotes intracellular Ca(2+) flux and reduces cAMP levels through a G protein-coupled receptor that signals via a Galpha(i/o) protein in THP1 cells. Toll-like receptor 4 stimulation reduces ubiquitin-binding sites, which enabled identification of four Galpha(i/o) PCRs as ubiquitin receptor candidates. Overexpression of candidate genes in HEK293 cells, gene silencing in THP1 cells, competition binding, and signaling studies with the CXCR4 agonist stromal cell-derived factor-1alpha (chemokine (CXC motif) ligand 12) and inhibitor AMD3100 identify CXCR4 as a functional ubiquitin receptor. Our finding uncovers a fundamentally new aspect of the role of ubiquitin in biology, has implications for the understanding of CXCR4-mediated events, and is expected to facilitate development of new therapeutic avenues for a variety of diseases.
Figures


 , biotin-ubiquitin (right ordinate; n = 3). ▵, FITC-SUMO1 (n = 5). ◇, FITC-SUMO2 (n = 5). □, FITC-ovalbumin (n = 3). ○, nonspecific binding (n = 6). Dashed line, specific binding curve. E, competition binding (1 min, 4 °C) curve for unlabeled ubiquitin (n = 6, ●) and ovalbumin (n = 3, □) with 1.16 μ
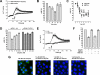
, biotin-ubiquitin (right ordinate; n = 3). ▵, FITC-SUMO1 (n = 5). ◇, FITC-SUMO2 (n = 5). □, FITC-ovalbumin (n = 3). ○, nonspecific binding (n = 6). Dashed line, specific binding curve. E, competition binding (1 min, 4 °C) curve for unlabeled ubiquitin (n = 6, ●) and ovalbumin (n = 3, □) with 1.16 μ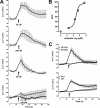
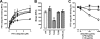


Similar articles
-
The CXC chemokine receptor 4 ligands ubiquitin and stromal cell-derived factor-1α function through distinct receptor interactions.J Biol Chem. 2011 Sep 23;286(38):33466-77. doi: 10.1074/jbc.M111.233742. Epub 2011 Jul 13. J Biol Chem. 2011. PMID: 21757744 Free PMC article.
-
CXC chemokine receptor 4 signaling upon co-activation with stromal cell-derived factor-1α and ubiquitin.Cytokine. 2014 Feb;65(2):121-5. doi: 10.1016/j.cyto.2013.12.008. Epub 2013 Dec 24. Cytokine. 2014. PMID: 24373940 Free PMC article.
-
Structural determinants of ubiquitin-CXC chemokine receptor 4 interaction.J Biol Chem. 2011 Dec 23;286(51):44145-44152. doi: 10.1074/jbc.M111.298505. Epub 2011 Oct 28. J Biol Chem. 2011. PMID: 22039044 Free PMC article.
-
Chemokine receptor CXCR4: role in gastrointestinal cancer.Crit Rev Oncol Hematol. 2013 Dec;88(3):696-705. doi: 10.1016/j.critrevonc.2013.08.005. Epub 2013 Aug 28. Crit Rev Oncol Hematol. 2013. PMID: 24120239 Review.
-
CXCL14 antagonizes the CXCL12-CXCR4 signaling axis.Biomol Concepts. 2014 May;5(2):167-73. doi: 10.1515/bmc-2014-0007. Biomol Concepts. 2014. PMID: 25372750 Review.
Cited by
-
Carcinoma cells misuse the host tissue damage response to invade the brain.Glia. 2013 Aug;61(8):1331-46. doi: 10.1002/glia.22518. Epub 2013 Jul 6. Glia. 2013. PMID: 23832647 Free PMC article.
-
Cardioprotective Potential of Exogenous Ubiquitin.Cardiovasc Drugs Ther. 2021 Dec;35(6):1227-1232. doi: 10.1007/s10557-020-07042-5. Epub 2020 Sep 10. Cardiovasc Drugs Ther. 2021. PMID: 32910339 Free PMC article. Review.
-
Modulation of the CXC chemokine receptor 4 agonist activity of ubiquitin through C-terminal protein modification.Biochemistry. 2013 Jun 18;52(24):4184-92. doi: 10.1021/bi400254f. Epub 2013 Jun 7. Biochemistry. 2013. PMID: 23697661 Free PMC article.
-
New paradigms in chemokine receptor signal transduction: Moving beyond the two-site model.Biochem Pharmacol. 2016 Aug 15;114:53-68. doi: 10.1016/j.bcp.2016.04.007. Epub 2016 Apr 19. Biochem Pharmacol. 2016. PMID: 27106080 Free PMC article. Review.
-
At the Bench: Pre-clinical evidence for multiple functions of CXCR4 in cancer.J Leukoc Biol. 2021 May;109(5):969-989. doi: 10.1002/JLB.2BT1018-715RR. Epub 2020 Oct 26. J Leukoc Biol. 2021. PMID: 33104270 Free PMC article. Review.
References
-
- Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Asseman C., Pancré V., Delanoye A., Capron A., Auriault C. (1994) J. Immunol. Methods 173, 93–101 - PubMed
-
- Takada K., Nasu H., Hibi N., Tsukada Y., Shibasaki T., Fujise K., Fujimuro M., Sawada H., Yokosawa H., Ohkawa K. (1997) Clin. Chem. 43, 1188–1195 - PubMed
-
- Takagi M., Yamauchi M., Toda G., Takada K., Hirakawa T., Ohkawa K. (1999) Alcohol. Clin. Exp. Res. 23, (Suppl. 4) 76S–80S - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

