FastTree 2--approximately maximum-likelihood trees for large alignments
- PMID: 20224823
- PMCID: PMC2835736
- DOI: 10.1371/journal.pone.0009490
FastTree 2--approximately maximum-likelihood trees for large alignments
Abstract
Background: We recently described FastTree, a tool for inferring phylogenies for alignments with up to hundreds of thousands of sequences. Here, we describe improvements to FastTree that improve its accuracy without sacrificing scalability.
Methodology/principal findings: Where FastTree 1 used nearest-neighbor interchanges (NNIs) and the minimum-evolution criterion to improve the tree, FastTree 2 adds minimum-evolution subtree-pruning-regrafting (SPRs) and maximum-likelihood NNIs. FastTree 2 uses heuristics to restrict the search for better trees and estimates a rate of evolution for each site (the "CAT" approximation). Nevertheless, for both simulated and genuine alignments, FastTree 2 is slightly more accurate than a standard implementation of maximum-likelihood NNIs (PhyML 3 with default settings). Although FastTree 2 is not quite as accurate as methods that use maximum-likelihood SPRs, most of the splits that disagree are poorly supported, and for large alignments, FastTree 2 is 100-1,000 times faster. FastTree 2 inferred a topology and likelihood-based local support values for 237,882 distinct 16S ribosomal RNAs on a desktop computer in 22 hours and 5.8 gigabytes of memory.
Conclusions/significance: FastTree 2 allows the inference of maximum-likelihood phylogenies for huge alignments. FastTree 2 is freely available at http://www.microbesonline.org/fasttree.
Conflict of interest statement
Figures
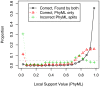
 + SPRs on simulated alignments with 250 protein sequences. We classified PhyML's splits as correct and found by both PhyML and FastTree, correct but missed by FastTree, or incorrect. We show the distribution of support values for each class. The right-most bin includes the strongly supported splits (0.95 to 1.0), and the gray dashed line shows the uniform distribution. The support values are PhyML's minimum of the approximate likelihood ratio test and SH-like , local supports.
+ SPRs on simulated alignments with 250 protein sequences. We classified PhyML's splits as correct and found by both PhyML and FastTree, correct but missed by FastTree, or incorrect. We show the distribution of support values for each class. The right-most bin includes the strongly supported splits (0.95 to 1.0), and the gray dashed line shows the uniform distribution. The support values are PhyML's minimum of the approximate likelihood ratio test and SH-like , local supports.
 axis). For RAxML with FastTree's (minimum-evolution) starting tree, we show the starting topology and RAxML's first two rounds of SPR moves.
axis). For RAxML with FastTree's (minimum-evolution) starting tree, we show the starting topology and RAxML's first two rounds of SPR moves.
Similar articles
-
FastTree: computing large minimum evolution trees with profiles instead of a distance matrix.Mol Biol Evol. 2009 Jul;26(7):1641-50. doi: 10.1093/molbev/msp077. Epub 2009 Apr 17. Mol Biol Evol. 2009. PMID: 19377059 Free PMC article.
-
RAxML and FastTree: comparing two methods for large-scale maximum likelihood phylogeny estimation.PLoS One. 2011;6(11):e27731. doi: 10.1371/journal.pone.0027731. Epub 2011 Nov 21. PLoS One. 2011. PMID: 22132132 Free PMC article.
-
morePhyML: improving the phylogenetic tree space exploration with PhyML 3.Mol Phylogenet Evol. 2011 Dec;61(3):944-8. doi: 10.1016/j.ympev.2011.08.029. Epub 2011 Sep 8. Mol Phylogenet Evol. 2011. PMID: 21925283
-
Very Fast Tree: speeding up the estimation of phylogenies for large alignments through parallelization and vectorization strategies.Bioinformatics. 2020 Nov 1;36(17):4658-4659. doi: 10.1093/bioinformatics/btaa582. Bioinformatics. 2020. PMID: 32573652
-
New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0.Syst Biol. 2010 May;59(3):307-21. doi: 10.1093/sysbio/syq010. Epub 2010 Mar 29. Syst Biol. 2010. PMID: 20525638
Cited by
-
Taxonomic Diversity, Predicted Metabolic Pathway, and Interaction Pattern of Bacterial Community in Sea Urchin Anthocidaris crassispina.Microorganisms. 2024 Oct 20;12(10):2094. doi: 10.3390/microorganisms12102094. Microorganisms. 2024. PMID: 39458402 Free PMC article.
-
Ethylene and epoxyethane metabolism in methanotrophic bacteria: comparative genomics and physiological studies using Methylohalobius crimeensis.Microb Genom. 2024 Oct;10(10):001306. doi: 10.1099/mgen.0.001306. Microb Genom. 2024. PMID: 39453690 Free PMC article.
-
Succeed to culture a novel lineage symbiotic bacterium of Mollicutes which widely found in arthropods intestine uncovers the potential double-edged sword ecological function.Front Microbiol. 2024 Oct 18;15:1458382. doi: 10.3389/fmicb.2024.1458382. eCollection 2024. Front Microbiol. 2024. PMID: 39493855 Free PMC article.
-
Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park.Biol Direct. 2013 Apr 22;8:9. doi: 10.1186/1745-6150-8-9. Biol Direct. 2013. PMID: 23607440 Free PMC article.
-
Upper airways microbiota in antibiotic-naïve wheezing and healthy infants from the tropics of rural Ecuador.PLoS One. 2012;7(10):e46803. doi: 10.1371/journal.pone.0046803. Epub 2012 Oct 5. PLoS One. 2012. PMID: 23071640 Free PMC article.
References
-
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. - PubMed
-
- Studier JA, Keppler KJ. A note on the neighbor-joining algorithm of Saitou and Nei. Mol Biol Evol. 1988;5:729–31. - PubMed
-
- Felsenstein J. Evolutionary trees from dna sequences: A maximum likelihood approach. J Mol Evol. 1981;17:368–376. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

