Assembly dynamics of PML nuclear bodies in living cells
- PMID: 20205709
- PMCID: PMC2854101
- DOI: 10.1186/1757-5036-3-3
Assembly dynamics of PML nuclear bodies in living cells
Abstract
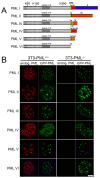
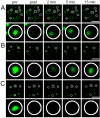
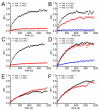
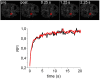
The mammalian cell nucleus contains a variety of organelles or nuclear bodies which contribute to key nuclear functions. Promyelocytic leukemia nuclear bodies (PML NBs) are involved in the regulation of apoptosis, antiviral responses, the DNA damage response and chromatin structure, but their precise biochemical function in these nuclear pathways is unknown. One strategy to tackle this problem is to assess the biophysical properties of the component parts of these macromolecular assemblies in living cells. In this study we determined PML NB assembly dynamics by live cell imaging, combined with mathematical modeling. For the first time, dynamics of PML body formation were measured in cells lacking endogenous PML. We show that all six human nuclear PML isoforms are able to form nuclear bodies in PML negative cells. All isoforms exhibit individual exchange rates at NBs in PML positive cells but PML I, II, III and IV are static at nuclear bodies in PML negative cells, suggesting that these isoforms require additional protein partners for efficient exchange. PML V turns over at PML Nbs very slowly supporting the idea of a structural function for this isoform. We also demonstrate that SUMOylation of PML at Lysine positions K160 and/or K490 are required for nuclear body formation in vivo.We propose a model in which the isoform specific residence times of PML provide both, structural stability to function as a scaffold and flexibility to attract specific nuclear proteins for efficient biochemical reactions at the surface of nuclear bodies.MCS code: 92C37.
Figures

Similar articles
-
Dynamics of component exchange at PML nuclear bodies.J Cell Sci. 2008 Aug 15;121(Pt 16):2731-43. doi: 10.1242/jcs.031922. Epub 2008 Jul 29. J Cell Sci. 2008. PMID: 18664490
-
C-terminal motifs in promyelocytic leukemia protein isoforms critically regulate PML nuclear body formation.J Cell Sci. 2017 Oct 15;130(20):3496-3506. doi: 10.1242/jcs.202879. Epub 2017 Aug 29. J Cell Sci. 2017. PMID: 28851805
-
The Human Cytomegalovirus IE1 Protein Antagonizes PML Nuclear Body-Mediated Intrinsic Immunity via the Inhibition of PML De Novo SUMOylation.J Virol. 2017 Jan 31;91(4):e02049-16. doi: 10.1128/JVI.02049-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27903803 Free PMC article.
-
Multimodal Light Microscopy Approaches to Reveal Structural and Functional Properties of Promyelocytic Leukemia Nuclear Bodies.Front Oncol. 2018 May 25;8:125. doi: 10.3389/fonc.2018.00125. eCollection 2018. Front Oncol. 2018. PMID: 29888200 Free PMC article. Review.
-
PML nuclear bodies and chromatin dynamics: catch me if you can!Nucleic Acids Res. 2020 Dec 2;48(21):11890-11912. doi: 10.1093/nar/gkaa828. Nucleic Acids Res. 2020. PMID: 33068409 Free PMC article. Review.
Cited by
-
PML isoforms IV and V contribute to adenovirus-mediated oncogenic transformation by functionally inhibiting the tumor-suppressor p53.Oncogene. 2016 Jan 7;35(1):69-82. doi: 10.1038/onc.2015.63. Epub 2015 Mar 16. Oncogene. 2016. PMID: 25772236
-
The Role of Non-Specific Interactions in Canonical and ALT-Associated PML-Bodies Formation and Dynamics.Int J Mol Sci. 2021 May 29;22(11):5821. doi: 10.3390/ijms22115821. Int J Mol Sci. 2021. PMID: 34072343 Free PMC article.
-
The p97/VCP segregase is essential for arsenic-induced degradation of PML and PML-RARA.J Cell Biol. 2023 Apr 3;222(4):e202201027. doi: 10.1083/jcb.202201027. Epub 2023 Feb 28. J Cell Biol. 2023. PMID: 36880596 Free PMC article.
-
Phase Separation in the Nucleus and at the Nuclear Periphery during Post-Mitotic Nuclear Envelope Reformation.Cells. 2022 May 25;11(11):1749. doi: 10.3390/cells11111749. Cells. 2022. PMID: 35681444 Free PMC article. Review.
-
Relationships between Cargo, Cell Penetrating Peptides and Cell Type for Uptake of Non-Covalent Complexes into Live Cells.Pharmaceuticals (Basel). 2013 Feb 6;6(2):184-203. doi: 10.3390/ph6020184. Pharmaceuticals (Basel). 2013. PMID: 24275947 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

