Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns
- PMID: 20203697
- PMCID: PMC2854594
- DOI: 10.1038/embor.2010.27
Differences in the DNA replication of unicellular eukaryotes and metazoans: known unknowns
Abstract
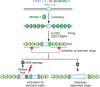
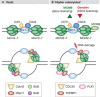
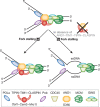
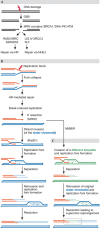
Although the basic mechanisms of DNA synthesis are conserved across species, there are differences between simple and complex organisms. In contrast to lower eukaryotes, replication origins in complex eukaryotes lack DNA sequence specificity, can be activated in response to stressful conditions and require poorly conserved factors for replication firing. The response to replication fork damage is monitored by conserved proteins, such as the TIPIN-TIM-CLASPIN complex. The absence of this complex induces severe effects on yeast replication, whereas in higher eukaryotes it is only crucial when the availability of replication origins is limiting. Finally, the dependence of DNA replication on homologous recombination proteins such as RAD51 and the MRE11-RAD50-NBS1 complex is also different; they are dispensable for yeast S-phase but essential for accurate DNA replication in metazoans under unchallenged conditions. The reasons for these differences are not yet understood. Here, we focus on some of these known unknowns of DNA replication.
Figures


Similar articles
-
Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors.J Mol Biol. 2007 Feb 9;366(1):36-52. doi: 10.1016/j.jmb.2006.10.097. Epub 2006 Nov 3. J Mol Biol. 2007. PMID: 17141802 Free PMC article.
-
ATM and the Mre11-Rad50-Nbs1 complex respond to nucleoside analogue-induced stalled replication forks and contribute to drug resistance.Cancer Res. 2008 Oct 1;68(19):7947-55. doi: 10.1158/0008-5472.CAN-08-0971. Cancer Res. 2008. PMID: 18829552 Free PMC article.
-
Separation of intra-S checkpoint protein contributions to DNA replication fork protection and genomic stability in normal human fibroblasts.Cell Cycle. 2013 Jan 15;12(2):332-45. doi: 10.4161/cc.23177. Epub 2012 Jan 15. Cell Cycle. 2013. PMID: 23255133 Free PMC article.
-
Surviving chromosome replication: the many roles of the S-phase checkpoint pathway.Philos Trans R Soc Lond B Biol Sci. 2011 Dec 27;366(1584):3554-61. doi: 10.1098/rstb.2011.0071. Philos Trans R Soc Lond B Biol Sci. 2011. PMID: 22084382 Free PMC article. Review.
-
Principles and concepts of DNA replication in bacteria, archaea, and eukarya.Cold Spring Harb Perspect Biol. 2013 Jul 1;5(7):a010108. doi: 10.1101/cshperspect.a010108. Cold Spring Harb Perspect Biol. 2013. PMID: 23818497 Free PMC article. Review.
Cited by
-
The cancer therapeutic potential of Chk1 inhibitors: how mechanistic studies impact on clinical trial design.Br J Clin Pharmacol. 2013 Sep;76(3):358-69. doi: 10.1111/bcp.12139. Br J Clin Pharmacol. 2013. PMID: 23593991 Free PMC article. Review.
-
Changes in soil taxonomic and functional diversity resulting from gamma irradiation.Sci Rep. 2019 May 27;9(1):7894. doi: 10.1038/s41598-019-44441-7. Sci Rep. 2019. PMID: 31133738 Free PMC article.
-
Balancing self-renewal against genome preservation in stem cells: How do they manage to have the cake and eat it too?Cell Mol Life Sci. 2016 May;73(9):1803-23. doi: 10.1007/s00018-016-2152-y. Epub 2016 Feb 17. Cell Mol Life Sci. 2016. PMID: 26886024 Free PMC article. Review.
-
Spatiotemporally different DNA repair systems participate in Epstein-Barr virus genome maturation.J Virol. 2011 Jul;85(13):6127-35. doi: 10.1128/JVI.00258-11. Epub 2011 Apr 13. J Virol. 2011. PMID: 21490093 Free PMC article.
-
Regulation of the cell division cycle in Trypanosoma brucei.Eukaryot Cell. 2012 Oct;11(10):1180-90. doi: 10.1128/EC.00145-12. Epub 2012 Aug 3. Eukaryot Cell. 2012. PMID: 22865501 Free PMC article. Review.
References
-
- Aylon Y, Kupiec M (2004) DSB repair: the yeast paradigm. DNA Repair (Amst) 3: 797–815 - PubMed
-
- Bell SP, Stillman B (1992) ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 - PubMed
-
- Branzei D, Foiani M (2005) The DNA damage response during DNA replication. Curr Opin Cell Biol 17: 568–575 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

