Hydrogen exchange mass spectrometry: what is it and what can it tell us?
- PMID: 20195578
- PMCID: PMC2868954
- DOI: 10.1007/s00216-010-3556-4
Hydrogen exchange mass spectrometry: what is it and what can it tell us?
Abstract
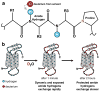
Proteins are undoubtedly some of the most essential molecules of life. While much is known about many proteins, some aspects still remain mysterious. One particularly important aspect of understanding proteins is determining how structure helps dictate function. Continued development and implementation of biophysical techniques that provide information about protein conformation and dynamics is essential. In this review, we discuss hydrogen exchange mass spectrometry and how this method can be used to learn about protein conformation and dynamics. The basic concepts of the method are described, the workflow illustrated, and a few examples of its application are provided.
Figures



Similar articles
-
Hydrogen exchange and mass spectrometry: A historical perspective.J Am Soc Mass Spectrom. 2006 Nov;17(11):1481-1489. doi: 10.1016/j.jasms.2006.06.006. Epub 2006 Jul 28. J Am Soc Mass Spectrom. 2006. PMID: 16876429 Free PMC article. Review.
-
Hydrogen Exchange Mass Spectrometry.Methods Enzymol. 2016;566:335-56. doi: 10.1016/bs.mie.2015.06.035. Epub 2015 Jul 27. Methods Enzymol. 2016. PMID: 26791986 Free PMC article.
-
Fragmentation hydrogen exchange mass spectrometry: a review of methodology and applications.Protein Expr Purif. 2012 Jul;84(1):19-37. doi: 10.1016/j.pep.2012.04.009. Epub 2012 Apr 23. Protein Expr Purif. 2012. PMID: 22554819 Review.
-
Mapping Protein-Ligand Interactions with Proteolytic Fragmentation, Hydrogen/Deuterium Exchange-Mass Spectrometry.Methods Enzymol. 2016;566:357-404. doi: 10.1016/bs.mie.2015.08.010. Epub 2015 Oct 23. Methods Enzymol. 2016. PMID: 26791987
-
Protein analysis by hydrogen exchange mass spectrometry.Annu Rev Biophys Biomol Struct. 2003;32:1-25. doi: 10.1146/annurev.biophys.32.110601.142417. Epub 2003 Feb 18. Annu Rev Biophys Biomol Struct. 2003. PMID: 12598366 Review.
Cited by
-
Structure and dynamic regulation of Abl kinases.J Biol Chem. 2013 Feb 22;288(8):5443-50. doi: 10.1074/jbc.R112.438382. Epub 2013 Jan 11. J Biol Chem. 2013. PMID: 23316053 Free PMC article. Review.
-
Minimizing back exchange in the hydrogen exchange-mass spectrometry experiment.J Am Soc Mass Spectrom. 2012 Dec;23(12):2132-9. doi: 10.1007/s13361-012-0476-x. Epub 2012 Sep 11. J Am Soc Mass Spectrom. 2012. PMID: 22965280 Free PMC article.
-
Characterizing monoclonal antibody structure by carboxyl group footprinting.MAbs. 2015;7(3):540-52. doi: 10.1080/19420862.2015.1023683. MAbs. 2015. PMID: 25933350 Free PMC article.
-
Cutting edge: Evidence for a dynamically driven T cell signaling mechanism.J Immunol. 2012 Jun 15;188(12):5819-23. doi: 10.4049/jimmunol.1200952. Epub 2012 May 18. J Immunol. 2012. PMID: 22611242 Free PMC article.
-
Membrane phospholipid bilayer as a determinant of monoacylglycerol lipase kinetic profile and conformational repertoire.Protein Sci. 2013 Jun;22(6):774-87. doi: 10.1002/pro.2257. Epub 2013 Apr 29. Protein Sci. 2013. PMID: 23553709 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

