Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity
- PMID: 20189389
- PMCID: PMC2910546
- DOI: 10.1016/j.cub.2009.12.062
Tubulin glutamylation regulates ciliary motility by altering inner dynein arm activity
Abstract
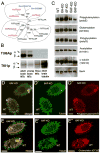
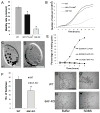
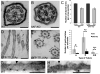
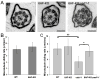
How microtubule-associated motor proteins are regulated is not well understood. A potential mechanism for spatial regulation of motor proteins is provided by posttranslational modifications of tubulin subunits that form patterns on microtubules. Glutamylation is a conserved tubulin modification [1] that is enriched in axonemes. The enzymes responsible for this posttranslational modification, glutamic acid ligases (E-ligases), belong to a family of proteins with a tubulin tyrosine ligase (TTL) homology domain (TTL-like or TTLL proteins) [2]. We show that in cilia of Tetrahymena, TTLL6 E-ligases generate glutamylation mainly on the B-tubule of outer doublet microtubules, the site of force production by ciliary dynein. Deletion of two TTLL6 paralogs caused severe deficiency in ciliary motility associated with abnormal waveform and reduced beat frequency. In isolated axonemes with a normal dynein arm composition, TTLL6 deficiency did not affect the rate of ATP-induced doublet microtubule sliding. Unexpectedly, the same TTLL6 deficiency increased the velocity of microtubule sliding in axonemes that also lack outer dynein arms, in which forces are generated by inner dynein arms. We conclude that tubulin glutamylation on the B-tubule inhibits the net force imposed on sliding doublet microtubules by inner dynein arms.
Figures
Similar articles
-
Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila.Eukaryot Cell. 2008 Aug;7(8):1362-72. doi: 10.1128/EC.00084-08. Epub 2008 Jun 27. Eukaryot Cell. 2008. PMID: 18586949 Free PMC article.
-
Glutamylation Regulates Transport, Specializes Function, and Sculpts the Structure of Cilia.Curr Biol. 2017 Nov 20;27(22):3430-3441.e6. doi: 10.1016/j.cub.2017.09.066. Epub 2017 Nov 9. Curr Biol. 2017. PMID: 29129530 Free PMC article.
-
Posttranslational Modifications of Tubulin and Cilia.Cold Spring Harb Perspect Biol. 2017 Jun 1;9(6):a028159. doi: 10.1101/cshperspect.a028159. Cold Spring Harb Perspect Biol. 2017. PMID: 28003186 Free PMC article. Review.
-
Tubulin glycylation controls ciliary motility through modulation of outer-arm dyneins.Mol Biol Cell. 2024 Jul 1;35(7):ar90. doi: 10.1091/mbc.E24-04-0154. Epub 2024 May 17. Mol Biol Cell. 2024. PMID: 38758663 Free PMC article.
-
Ciliary Motility: Regulation of Axonemal Dynein Motors.Cold Spring Harb Perspect Biol. 2017 Aug 1;9(8):a018325. doi: 10.1101/cshperspect.a018325. Cold Spring Harb Perspect Biol. 2017. PMID: 28765157 Free PMC article. Review.
Cited by
-
The motility of axonemal dynein is regulated by the tubulin code.Biophys J. 2014 Dec 16;107(12):2872-2880. doi: 10.1016/j.bpj.2014.10.061. Biophys J. 2014. PMID: 25658008 Free PMC article.
-
The Emerging Roles of Axonemal Glutamylation in Regulation of Cilia Architecture and Functions.Front Cell Dev Biol. 2021 Mar 4;9:622302. doi: 10.3389/fcell.2021.622302. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33748109 Free PMC article. Review.
-
Proximity Mapping of CCP6 Reveals Its Association with Centrosome Organization and Cilium Assembly.Int J Mol Sci. 2023 Jan 9;24(2):1273. doi: 10.3390/ijms24021273. Int J Mol Sci. 2023. PMID: 36674791 Free PMC article.
-
Deciphering the Tubulin Language: Molecular Determinants and Readout Mechanisms of the Tubulin Code in Neurons.Int J Mol Sci. 2023 Feb 1;24(3):2781. doi: 10.3390/ijms24032781. Int J Mol Sci. 2023. PMID: 36769099 Free PMC article. Review.
-
Proteins that control the geometry of microtubules at the ends of cilia.J Cell Biol. 2018 Dec 3;217(12):4298-4313. doi: 10.1083/jcb.201804141. Epub 2018 Sep 14. J Cell Biol. 2018. PMID: 30217954 Free PMC article.
References
-
- Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. - PubMed
-
- Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–1762. - PubMed
-
- Regnard C, Fesquet D, Janke C, Boucher D, Desbruyeres E, Koulakoff A, Insina C, Travo P, Edde B. Characterisation of PGs1, a subunit of a protein complex co-purifying with tubulin polyglutamylase. J Cell Sci. 2003;116:4181–4190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

