The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans
- PMID: 20176573
- PMCID: PMC2887956
- DOI: 10.1093/nar/gkq083
The microRNA miR-124 controls gene expression in the sensory nervous system of Caenorhabditis elegans
Abstract
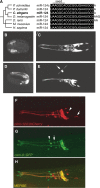
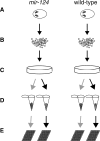
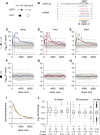
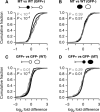
miR-124 is a highly conserved microRNA (miRNA) whose in vivo function is poorly understood. Here, we identify miR-124 targets based on the analysis of the first mir-124 mutant in any organism. We find that miR-124 is expressed in many sensory neurons in Caenorhabditis elegans and onset of expression coincides with neuronal morphogenesis. We analyzed the transcriptome of miR-124 expressing and nonexpressing cells from wild-type and mir-124 mutants. We observe that many targets are co-expressed with and actively repressed by miR-124. These targets are expressed at reduced relative levels in sensory neurons compared to the rest of the animal. Our data from mir-124 mutant animals show that this effect is due to a large extent to the activity of miR-124. Genes with nonconserved target sites show reduced absolute expression levels in sensory neurons. In contrast, absolute expression levels of genes with conserved sites are comparable to control genes, suggesting a tuning function for many of these targets. We conclude that miR-124 contributes to defining cell-type-specific gene activity by repressing a diverse set of co-expressed genes.
Figures


Similar articles
-
Neuron type-specific miRNA represses two broadly expressed genes to modulate an avoidance behavior in C. elegans.Genes Dev. 2016 Sep 15;30(18):2042-2047. doi: 10.1101/gad.287904.116. Epub 2016 Sep 29. Genes Dev. 2016. PMID: 27688400 Free PMC article.
-
A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans.Nature. 2003 Dec 18;426(6968):845-9. doi: 10.1038/nature02255. Epub 2003 Dec 14. Nature. 2003. PMID: 14685240
-
IgCAMs redundantly control axon navigation in Caenorhabditis elegans.Neural Dev. 2009 Apr 2;4:13. doi: 10.1186/1749-8104-4-13. Neural Dev. 2009. PMID: 19341471 Free PMC article.
-
Roles of microRNAs in the Caenorhabditis elegans nervous system.J Genet Genomics. 2013 Sep 20;40(9):445-52. doi: 10.1016/j.jgg.2013.07.002. Epub 2013 Aug 7. J Genet Genomics. 2013. PMID: 24053946 Review.
-
The microRNAs of Caenorhabditis elegans.Semin Cell Dev Biol. 2010 Sep;21(7):728-37. doi: 10.1016/j.semcdb.2010.07.001. Epub 2010 Jul 15. Semin Cell Dev Biol. 2010. PMID: 20637886 Review.
Cited by
-
Predicting microRNA targeting efficacy in Drosophila.Genome Biol. 2018 Oct 4;19(1):152. doi: 10.1186/s13059-018-1504-3. Genome Biol. 2018. PMID: 30286781 Free PMC article.
-
Functional shifts in insect microRNA evolution.Genome Biol Evol. 2010;2:686-96. doi: 10.1093/gbe/evq053. Epub 2010 Sep 3. Genome Biol Evol. 2010. PMID: 20817720 Free PMC article.
-
STAT3 is involved in miR-124-mediated suppressive effects on esophageal cancer cells.BMC Cancer. 2015 Apr 19;15:306. doi: 10.1186/s12885-015-1303-0. BMC Cancer. 2015. PMID: 25928665 Free PMC article.
-
Cell-type specific sequencing of microRNAs from complex animal tissues.Nat Methods. 2018 Apr;15(4):283-289. doi: 10.1038/nmeth.4610. Epub 2018 Feb 26. Nat Methods. 2018. PMID: 29481550 Free PMC article.
-
Overexpression of microRNA-124 promotes the neuronal differentiation of bone marrow-derived mesenchymal stem cells.Neural Regen Res. 2014 Jun 15;9(12):1241-8. doi: 10.4103/1673-5374.135333. Neural Regen Res. 2014. PMID: 25206789 Free PMC article.
References
-
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. - PubMed
-
- Arazi T, Talmor-Neiman M, Stav R, Riese M, Huijser P, Baulcombe DC. Cloning and characterization of micro-RNAs from moss. Plant J. 2005;43:837–848. - PubMed
-
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

