A new nuclear function of the Entamoeba histolytica glycolytic enzyme enolase: the metabolic regulation of cytosine-5 methyltransferase 2 (Dnmt2) activity
- PMID: 20174608
- PMCID: PMC2824750
- DOI: 10.1371/journal.ppat.1000775
A new nuclear function of the Entamoeba histolytica glycolytic enzyme enolase: the metabolic regulation of cytosine-5 methyltransferase 2 (Dnmt2) activity
Abstract
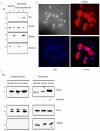
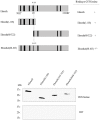
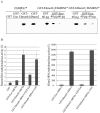
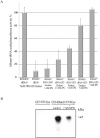
Cytosine-5 methyltransferases of the Dnmt2 family function as DNA and tRNA methyltransferases. Insight into the role and biological significance of Dnmt2 is greatly hampered by a lack of knowledge about its protein interactions. In this report, we address the subject of protein interaction by identifying enolase through a yeast two-hybrid screen as a Dnmt2-binding protein. Enolase, which is known to catalyze the conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP), was shown to have both a cytoplasmatic and a nuclear localization in the parasite Entamoeba histolytica. We discovered that enolase acts as a Dnmt2 inhibitor. This unexpected inhibitory activity was antagonized by 2-PG, which suggests that glucose metabolism controls the non-glycolytic function of enolase. Interestingly, glucose starvation drives enolase to accumulate within the nucleus, which in turn leads to the formation of additional enolase-E.histolytica DNMT2 homolog (Ehmeth) complex, and to a significant reduction of the tRNA(Asp) methylation in the parasite. The crucial role of enolase as a Dnmt2 inhibitor was also demonstrated in E.histolytica expressing a nuclear localization signal (NLS)-fused-enolase. These results establish enolase as the first Dnmt2 interacting protein, and highlight an unexpected role of a glycolytic enzyme in the modulation of Dnmt2 activity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
The Entamoeba histolytica Dnmt2 homolog (Ehmeth) confers resistance to nitrosative stress.Eukaryot Cell. 2014 Apr;13(4):494-503. doi: 10.1128/EC.00031-14. Epub 2014 Feb 21. Eukaryot Cell. 2014. PMID: 24562908 Free PMC article.
-
In vitro tRNA methylation assay with the Entamoeba histolytica DNA and tRNA methyltransferase Dnmt2 (Ehmeth) enzyme.J Vis Exp. 2010 Oct 19;(44):2390. doi: 10.3791/2390. J Vis Exp. 2010. PMID: 21048666 Free PMC article.
-
Characterization of cytosine methylated regions and 5-cytosine DNA methyltransferase (Ehmeth) in the protozoan parasite Entamoeba histolytica.Nucleic Acids Res. 2004 Jan 9;32(1):287-97. doi: 10.1093/nar/gkh161. Print 2004. Nucleic Acids Res. 2004. PMID: 14715927 Free PMC article.
-
Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification.Biomolecules. 2017 Feb 10;7(1):14. doi: 10.3390/biom7010014. Biomolecules. 2017. PMID: 28208632 Free PMC article. Review.
-
Solving the Dnmt2 enigma.Chromosoma. 2010 Feb;119(1):35-40. doi: 10.1007/s00412-009-0240-6. Chromosoma. 2010. PMID: 19730874 Review.
Cited by
-
New substrates and determinants for tRNA recognition of RNA methyltransferase DNMT2/TRDMT1.RNA Biol. 2021 Dec;18(12):2531-2545. doi: 10.1080/15476286.2021.1930756. Epub 2021 Jun 10. RNA Biol. 2021. PMID: 34110975 Free PMC article.
-
The identification and characterization of non-coding and coding RNAs and their modified nucleosides by mass spectrometry.RNA Biol. 2014;11(12):1568-85. doi: 10.4161/15476286.2014.992280. RNA Biol. 2014. PMID: 25616408 Free PMC article. Review.
-
Transferrin: Endocytosis and Cell Signaling in Parasitic Protozoa.Biomed Res Int. 2015;2015:641392. doi: 10.1155/2015/641392. Epub 2015 May 18. Biomed Res Int. 2015. PMID: 26090431 Free PMC article. Review.
-
Are Metabolites From the Gut Microbiota Capable of Regulating Epigenetic Mechanisms in the Human Parasite Entamoeba histolytica?Front Cell Dev Biol. 2022 Mar 1;10:841586. doi: 10.3389/fcell.2022.841586. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35300430 Free PMC article. Review.
-
Structure analysis of Entamoeba histolytica DNMT2 (EhMeth).PLoS One. 2012;7(6):e38728. doi: 10.1371/journal.pone.0038728. Epub 2012 Jun 21. PLoS One. 2012. PMID: 22737219 Free PMC article.
References
-
- Spada F, Rothbauer U, Zolghadr K, Schermelleh L, Leonhardt H. Regulation of DNA methyltransferase 1. Adv Enzyme Regul. 2006;46:224–234. - PubMed
-
- Jeltsch A. Molecular enzymology of mammalian DNA methyltransferases. Curr Top Microbiol Immunol. 2006;301:203–225. - PubMed
-
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science. 2006;311:395–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

