Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease
- PMID: 20174605
- PMCID: PMC2824747
- DOI: 10.1371/journal.pgen.1000851
Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease
Abstract
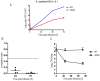
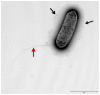
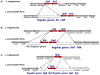
Legionella pneumophila and L. longbeachae are two species of a large genus of bacteria that are ubiquitous in nature. L. pneumophila is mainly found in natural and artificial water circuits while L. longbeachae is mainly present in soil. Under the appropriate conditions both species are human pathogens, capable of causing a severe form of pneumonia termed Legionnaires' disease. Here we report the sequencing and analysis of four L. longbeachae genomes, one complete genome sequence of L. longbeachae strain NSW150 serogroup (Sg) 1, and three draft genome sequences another belonging to Sg1 and two to Sg2. The genome organization and gene content of the four L. longbeachae genomes are highly conserved, indicating strong pressure for niche adaptation. Analysis and comparison of L. longbeachae strain NSW150 with L. pneumophila revealed common but also unexpected features specific to this pathogen. The interaction with host cells shows distinct features from L. pneumophila, as L. longbeachae possesses a unique repertoire of putative Dot/Icm type IV secretion system substrates, eukaryotic-like and eukaryotic domain proteins, and encodes additional secretion systems. However, analysis of the ability of a dotA mutant of L. longbeachae NSW150 to replicate in the Acanthamoeba castellanii and in a mouse lung infection model showed that the Dot/Icm type IV secretion system is also essential for the virulence of L. longbeachae. In contrast to L. pneumophila, L. longbeachae does not encode flagella, thereby providing a possible explanation for differences in mouse susceptibility to infection between the two pathogens. Furthermore, transcriptome analysis revealed that L. longbeachae has a less pronounced biphasic life cycle as compared to L. pneumophila, and genome analysis and electron microscopy suggested that L. longbeachae is encapsulated. These species-specific differences may account for the different environmental niches and disease epidemiology of these two Legionella species.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Dot/Icm Effector Translocation by Legionella longbeachae Creates a Replicative Vacuole Similar to That of Legionella pneumophila despite Translocation of Distinct Effector Repertoires.Infect Immun. 2015 Oct;83(10):4081-92. doi: 10.1128/IAI.00461-15. Epub 2015 Jul 27. Infect Immun. 2015. PMID: 26216429 Free PMC article.
-
Virulence factors encoded by Legionella longbeachae identified on the basis of the genome sequence analysis of clinical isolate D-4968.J Bacteriol. 2010 Feb;192(4):1030-44. doi: 10.1128/JB.01272-09. Epub 2009 Dec 11. J Bacteriol. 2010. PMID: 20008069 Free PMC article.
-
Genetic susceptibility and caspase activation in mouse and human macrophages are distinct for Legionella longbeachae and L. pneumophila.Infect Immun. 2007 Apr;75(4):1933-45. doi: 10.1128/IAI.00025-07. Epub 2007 Jan 29. Infect Immun. 2007. PMID: 17261610 Free PMC article.
-
Legionella pneumophila adaptation to intracellular life and the host response: clues from genomics and transcriptomics.FEBS Lett. 2007 Jun 19;581(15):2829-38. doi: 10.1016/j.febslet.2007.05.026. Epub 2007 May 21. FEBS Lett. 2007. PMID: 17531986 Review.
-
Acanthamoeba and Dictyostelium as Cellular Models for Legionella Infection.Front Cell Infect Microbiol. 2018 Mar 2;8:61. doi: 10.3389/fcimb.2018.00061. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29552544 Free PMC article. Review.
Cited by
-
Bacterial communities of Beijing surface waters as revealed by 454 pyrosequencing of the 16S rRNA gene.Environ Sci Pollut Res Int. 2015 Aug;22(16):12605-14. doi: 10.1007/s11356-015-4534-3. Epub 2015 Apr 26. Environ Sci Pollut Res Int. 2015. PMID: 25911286
-
Detection of novel Chlamydiae and Legionellales from human nasal samples of healthy volunteers.Folia Microbiol (Praha). 2015 Jul;60(4):325-34. doi: 10.1007/s12223-015-0378-y. Epub 2015 Feb 20. Folia Microbiol (Praha). 2015. PMID: 25697709
-
Amoebae as training grounds for microbial pathogens.mBio. 2024 Aug 14;15(8):e0082724. doi: 10.1128/mbio.00827-24. Epub 2024 Jul 8. mBio. 2024. PMID: 38975782 Free PMC article. Review.
-
The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments.Environ Microbiol. 2014 Feb;16(2):359-81. doi: 10.1111/1462-2920.12223. Epub 2013 Aug 19. Environ Microbiol. 2014. PMID: 23957615 Free PMC article.
-
Intra-amoeba multiplication induces chemotaxis and biofilm colonization and formation for Legionella.PLoS One. 2013 Oct 24;8(10):e77875. doi: 10.1371/journal.pone.0077875. eCollection 2013. PLoS One. 2013. PMID: 24205008 Free PMC article.
References
-
- Yu VL, Plouffe JF, Pastoris MC, Stout JE, Schousboe M, et al. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J Infect Dis. 2002;186:127–128. - PubMed
-
- Phares CR, Wangroongsarb P, Chantra S, Paveenkitiporn W, Tondella ML, et al. Epidemiology of severe pneumonia caused by Legionella longbeachae, Mycoplasma pneumoniae, and Chlamydia pneumoniae: 1-year, population-based surveillance for severe pneumonia in Thailand. Clin Infect Dis. 2007;45:e147–155. - PubMed
-
- Cameron S, Roder D, Walker C, Feldheim J. Epidemiological characteristics of Legionella infection in South Australia: implications for disease control. Aust N Z J Med. 1991;21:65–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

