Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates
- PMID: 20164237
- PMCID: PMC2863767
- DOI: 10.1128/JVI.02198-09
Mechanistic consequences of hnRNP C binding to both RNA termini of poliovirus negative-strand RNA intermediates
Abstract
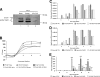
The poliovirus 3' noncoding region (3' NCR) is necessary for efficient virus replication. A poliovirus mutant, PVDelta3'NCR, with a deletion of the entire 3' NCR, yielded a virus that was capable of synthesizing viral RNA, albeit with a replication defect caused by deficient positive-strand RNA synthesis compared to wild-type virus. We detected multiple ribonucleoprotein (RNP) complexes in extracts from poliovirus-infected HeLa cells formed with a probe corresponding to the 5' end of poliovirus negative-strand RNA (the complement of the genomic 3' NCR), and the levels of these RNP complexes increased during the course of viral infection. Previous studies have identified RNP complexes formed with the 3' end of poliovirus negative-strand RNA, including one that contains a 36-kDa protein later identified as heterogeneous nuclear ribonucleoprotein C (hnRNP C). We report here that the 5' end of poliovirus negative-strand RNA is capable of interacting with endogenous hnRNP C, as well as with poliovirus nonstructural proteins. Further, we demonstrate that the addition of recombinant purified hnRNP C proteins can stimulate virus RNA synthesis in vitro and that depletion of hnRNP C proteins in cultured cells results in decreased virus yields and a correspondingly diminished accumulation of positive-strand RNAs. We propose that the association of hnRNP C with poliovirus negative-strand termini acts to stabilize or otherwise promote efficient positive-strand RNA synthesis.
Figures


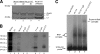

Similar articles
-
Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes.J Virol. 2005 Mar;79(6):3254-66. doi: 10.1128/JVI.79.6.3254-3266.2005. J Virol. 2005. PMID: 15731220 Free PMC article.
-
Delayed kinetics of poliovirus RNA synthesis in a human cell line with reduced levels of hnRNP C proteins.Virology. 2010 May 10;400(2):240-7. doi: 10.1016/j.virol.2010.01.031. Epub 2010 Mar 1. Virology. 2010. PMID: 20189623 Free PMC article.
-
An authentic 3' noncoding region is necessary for efficient poliovirus replication.J Virol. 2005 Sep;79(18):11962-73. doi: 10.1128/JVI.79.18.11962-11973.2005. J Virol. 2005. PMID: 16140772 Free PMC article.
-
Coxsackievirus B RNA replication: lessons from poliovirus.Curr Top Microbiol Immunol. 2008;323:89-121. doi: 10.1007/978-3-540-75546-3_5. Curr Top Microbiol Immunol. 2008. PMID: 18357767 Review.
-
Common replication strategies emerging from the study of diverse groups of positive-strand RNA viruses.Arch Virol Suppl. 1994;9:181-94. doi: 10.1007/978-3-7091-9326-6_18. Arch Virol Suppl. 1994. PMID: 8032249 Review.
Cited by
-
Non-template functions of viral RNA in picornavirus replication.Curr Opin Virol. 2011 Nov;1(5):339-46. doi: 10.1016/j.coviro.2011.09.005. Curr Opin Virol. 2011. PMID: 22140418 Free PMC article. Review.
-
Role of human heterogeneous nuclear ribonucleoprotein C1/C2 in dengue virus replication.Virol J. 2015 Feb 6;12:14. doi: 10.1186/s12985-014-0219-7. Virol J. 2015. PMID: 25890165 Free PMC article.
-
Making of viral replication organelles by remodeling interior membranes.Viruses. 2010 Nov;2(11):2436-42. doi: 10.3390/v2112436. Epub 2010 Nov 5. Viruses. 2010. PMID: 21994625 Free PMC article.
-
Early Emergence of 5' Terminally Deleted Coxsackievirus-B3 RNA Forms Is Associated with Acute and Persistent Infections in Mouse Target Tissues.Vaccines (Basel). 2022 Jul 28;10(8):1203. doi: 10.3390/vaccines10081203. Vaccines (Basel). 2022. PMID: 36016091 Free PMC article.
-
Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses.Virology. 2015 May;479-480:457-74. doi: 10.1016/j.virol.2015.03.001. Epub 2015 Mar 26. Virology. 2015. PMID: 25818028 Free PMC article. Review.
References
-
- Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. - PubMed
-
- Banerjee, R., W. Tsai, W. Kim, and A. Dasgupta. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 280:41-51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

