Role of calcineurin, hnRNPA2 and Akt in mitochondrial respiratory stress-mediated transcription activation of nuclear gene targets
- PMID: 20153290
- PMCID: PMC2891149
- DOI: 10.1016/j.bbabio.2010.02.008
Role of calcineurin, hnRNPA2 and Akt in mitochondrial respiratory stress-mediated transcription activation of nuclear gene targets
Abstract
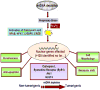
Pathophysiological conditions causing mitochondrial dysfunction and altered transmembrane potential (psim) initiate a mitochondrial respiratory stress response, also known as mitochondrial retrograde response, in a variety of mammalian cells. An increase in the cytosolic Ca2+ [Ca2+]c as part of this signaling cascade activates Ca2+ responsive phosphatase, calcineurin (Cn). Activation of IGF1R accompanied by increased glycolysis, invasiveness, and resistance to apoptosis is a phenotypic hallmark of C2C12 skeletal muscle cells subjected to this stress. The signaling is associated with activation and increased nuclear translocation of a number of transcription factors including a novel NFkappaB (cRel:p50) pathway, NFAT, CREB and C/EBPdelta. This culminates in the upregulation of a number of nuclear genes including Cathepsin L, RyR1, Glut4 and Akt1. We observed that stress regulated transcription activation of nuclear genes involves a cooperative interplay between NFkappaB (cRel:p50), C/EBPdelta, CREB, and NFAT. Our results show that the functional synergy of these factors requires the stress-activated heterogeneous nuclear ribonucleoprotein, hnRNPA2 as a transcriptional coactivator. We report here that mitochondrial stress leads to induced expression and activation of serine threonine kinase Akt1. Interestingly, we observe that Akt1 phosphorylates hnRNPA2 under mitochondrial stress conditions, which is a crucial step for the recruitment of this coactivator to the stress target promoters and culmination in mitochondrial stress-mediated transcription activation of target genes. We propose that mitochondrial stress plays an important role in tumor progression and emergence of invasive phenotypes.
Copyright © 2010. Published by Elsevier B.V.
Figures
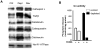





Similar articles
-
Activation of Akt is essential for the propagation of mitochondrial respiratory stress signaling and activation of the transcriptional coactivator heterogeneous ribonucleoprotein A2.Mol Biol Cell. 2010 Oct 15;21(20):3578-89. doi: 10.1091/mbc.E10-03-0192. Epub 2010 Aug 18. Mol Biol Cell. 2010. PMID: 20719961 Free PMC article.
-
Heterogeneous nuclear ribonucleoprotein A2 is a common transcriptional coactivator in the nuclear transcription response to mitochondrial respiratory stress.Mol Biol Cell. 2009 Sep;20(18):4107-19. doi: 10.1091/mbc.e09-04-0296. Epub 2009 Jul 29. Mol Biol Cell. 2009. PMID: 19641020 Free PMC article.
-
Enhanced osteoclastogenesis by mitochondrial retrograde signaling through transcriptional activation of the cathepsin K gene.Ann N Y Acad Sci. 2016 Jan;1364(1):52-61. doi: 10.1111/nyas.12709. Epub 2015 Mar 18. Ann N Y Acad Sci. 2016. PMID: 25800988 Free PMC article.
-
A systematic review of p53 regulation of oxidative stress in skeletal muscle.Redox Rep. 2018 Dec;23(1):100-117. doi: 10.1080/13510002.2017.1416773. Epub 2018 Jan 3. Redox Rep. 2018. PMID: 29298131 Free PMC article. Review.
-
Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics.Mitochondrion. 2013 Nov;13(6):577-91. doi: 10.1016/j.mito.2013.08.007. Epub 2013 Sep 1. Mitochondrion. 2013. PMID: 24004957 Free PMC article. Review.
Cited by
-
Mitochondria and cancer.Nat Rev Cancer. 2012 Oct;12(10):685-98. doi: 10.1038/nrc3365. Nat Rev Cancer. 2012. PMID: 23001348 Free PMC article. Review.
-
Absence of IκBβ/NFκB signaling does not attenuate acetaminophen-induced hepatic injury.Anat Rec (Hoboken). 2022 Nov 25:10.1002/ar.25126. doi: 10.1002/ar.25126. Online ahead of print. Anat Rec (Hoboken). 2022. PMID: 36426684 Free PMC article.
-
Mitochondrial retrograde signaling induces epithelial-mesenchymal transition and generates breast cancer stem cells.Oncogene. 2014 Nov 6;33(45):5238-50. doi: 10.1038/onc.2013.467. Epub 2013 Nov 4. Oncogene. 2014. PMID: 24186204 Free PMC article.
-
Roles of Mitochondrial DNA Changes on Cancer Initiation and Progression.Cell Biol (Henderson, NV). 2012 Oct 25;1(2):109. doi: 10.4172/2324-9293.1000e109. Cell Biol (Henderson, NV). 2012. PMID: 24319697 Free PMC article. No abstract available.
-
Mitochondrial Dysfunction: A Novel Potential Driver of Epithelial-to-Mesenchymal Transition in Cancer.Front Oncol. 2017 Dec 1;7:295. doi: 10.3389/fonc.2017.00295. eCollection 2017. Front Oncol. 2017. PMID: 29250487 Free PMC article. Review.
References
-
- Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 1999;18:522–533. - PMC - PubMed
-
- Murphy BJ, Robin ED, Tapper DP, Wong RJ, Clayton DA. Hypoxic coordinate regulation of mitochondrial enzymes in mammalian cells. Science. 1984;223:707–709. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

