Methods in mammalian autophagy research
- PMID: 20144757
- PMCID: PMC2852113
- DOI: 10.1016/j.cell.2010.01.028
Methods in mammalian autophagy research
Abstract
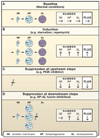
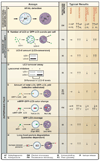
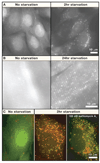
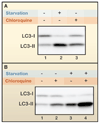
Autophagy has been implicated in many physiological and pathological processes. Accordingly, there is a growing scientific need to accurately identify, quantify, and manipulate the process of autophagy. However, as autophagy involves dynamic and complicated processes, it is often analyzed incorrectly. In this Primer, we discuss methods to monitor autophagy and to modulate autophagic activity, with a primary focus on mammalian macroautophagy.
2010 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Methods for Detection of Autophagy in Mammalian Cells.Methods Mol Biol. 2019;2045:245-258. doi: 10.1007/7651_2018_190. Methods Mol Biol. 2019. PMID: 30242567
-
Macroautophagy signaling and regulation.Curr Top Microbiol Immunol. 2009;335:33-70. doi: 10.1007/978-3-642-00302-8_2. Curr Top Microbiol Immunol. 2009. PMID: 19802559 Review.
-
Measuring macroautophagy in S. cerevisiae: autophagic body accumulation and total protein turnover.Methods Enzymol. 2008;451:57-66. doi: 10.1016/S0076-6879(08)03205-9. Methods Enzymol. 2008. PMID: 19185713
-
Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer's disease.J Neurosci. 2008 Jul 2;28(27):6926-37. doi: 10.1523/JNEUROSCI.0800-08.2008. J Neurosci. 2008. PMID: 18596167 Free PMC article.
-
Autophagosome formation and molecular mechanism of autophagy.Antioxid Redox Signal. 2011 Jun;14(11):2201-14. doi: 10.1089/ars.2010.3482. Epub 2010 Dec 4. Antioxid Redox Signal. 2011. PMID: 20712405 Review.
Cited by
-
Stigmasterol Simultaneously Induces Apoptosis and Protective Autophagy by Inhibiting Akt/mTOR Pathway in Gastric Cancer Cells.Front Oncol. 2021 Feb 23;11:629008. doi: 10.3389/fonc.2021.629008. eCollection 2021. Front Oncol. 2021. PMID: 33708631 Free PMC article.
-
Identification and Expression Analysis of the Solanum tuberosum StATG8 Family Associated with the WRKY Transcription Factor.Plants (Basel). 2022 Oct 26;11(21):2858. doi: 10.3390/plants11212858. Plants (Basel). 2022. PMID: 36365311 Free PMC article.
-
Virus recognition by Toll-7 activates antiviral autophagy in Drosophila.Immunity. 2012 Apr 20;36(4):658-67. doi: 10.1016/j.immuni.2012.03.003. Epub 2012 Mar 29. Immunity. 2012. PMID: 22464169 Free PMC article.
-
Placental extracellular vesicles and pre-eclampsia.Am J Reprod Immunol. 2021 Feb;85(2):e13297. doi: 10.1111/aji.13297. Epub 2020 Jul 18. Am J Reprod Immunol. 2021. PMID: 32619308 Free PMC article. Review.
-
Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway.Mol Neurodegener. 2012 Jan 9;7:2. doi: 10.1186/1750-1326-7-2. Mol Neurodegener. 2012. PMID: 22230652 Free PMC article.
References
-
- Bampton ETW, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autopha-gosome formation to fusion with endo/lysosomes. Autophagy. 2005;1:23–36. - PubMed
-
- Blommaart EFC, Krause U, Schellens JPM, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997;243:240–246. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

